Chapter: 11th Microbiology : Chapter 14 : Microbial Genetics
DNA Replication
DNA Replication
DNA is a marvelous device for the stable storage of genetic
information. DNA replication is a process in which copies of DNA molecules are
faithfully made. Here double stranded DNA molecule is copied to produce two identical
dsDNA molecules. Replication is an essential process because whenever a cell
divides, the two new daughter cells must contain the same genetic information
in DNA as parent cell. DNA replication occurs during the S (DNA synthesis)
phase and precedes cell division.
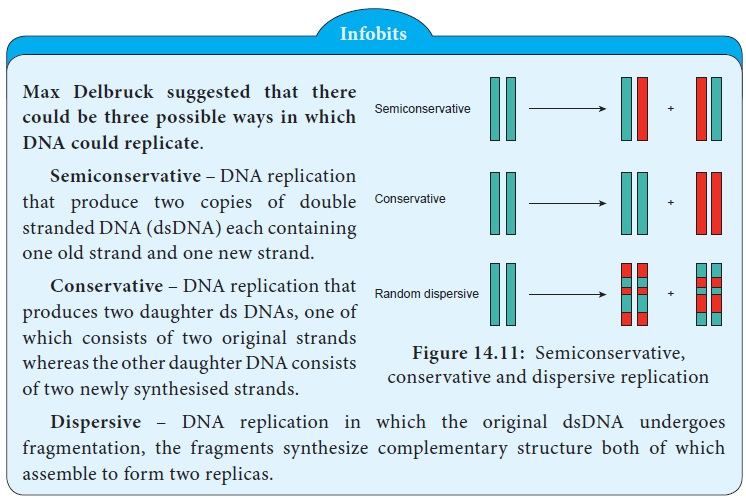
Watson and Crick proposed the hypothesis of semiconservative
replication. According to them each DNA strand serves as a template for the
synthesis of a new strand, producing two new DNA molecules each with one old
strand and one new strand. (Figure 14.11).
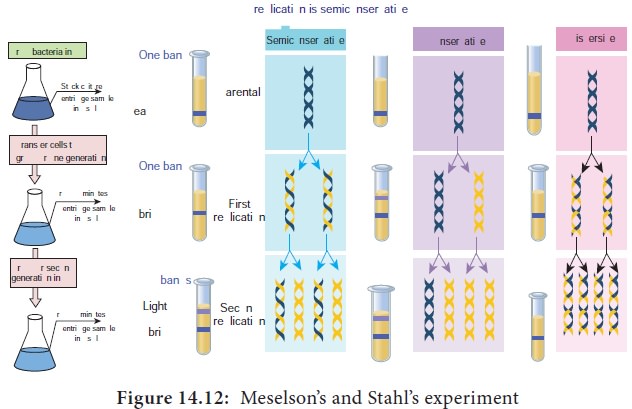
Meselson and Stahl’s Experiment
Mathew Meselson and Franklin Stahl in 1957 gave experimental evidence for semiconservative replica-tion process.
Steps
1.
E.coli cells were grown for
many generations in a medium
containing radioactive isotope (heavy isotope) of nitrogen source 15N.
2.
After many generations all nitrogen containing molecules in E.coli cells, including nitrogen bases
of DNA contained 15N.
3.
The cells labeled with 15N were then transferred to a
medium containing only 14N (light isotope). Hence all subsequent DNA
synthesised during replication contained 14N.
4.
Cell samples were removed at periodic time intervals from the
growth medium.
5.
From each of the above samples, DNA was isolated and subjected
to density gradient centrifugation (Caesium chloride (CsCl) centrifugation).
Expected results
·
The heavy isotope 15N containing DNA, will reach
equilibrium in a gradient point closer to the bottom of the tube.
·
14N containing DNA will reach equilibrium at a gradient point
closer to the surface of the tube.
Observed Results
·
After first generation the isolated DNA occupied an intermediate
density band.
·
After second generation two bands were observed, one at intermediate
density and the other at lighter density corresponding to the 14N
position in the gradient.
These experimental results and other experiments repeated by
Meselson and Stahl with prokaryotes suggested that semiconservative mechanism
of replication is universal (Figure 14.12).
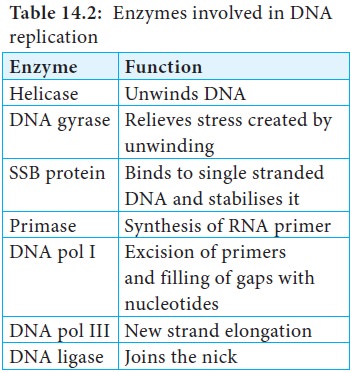
Enzymes Involved in DNA Replication
DNA replication in E.coli
requires many enzymes and proteins, each performing a specific task. The entire
complex is called the DNA replicase system or replisome. The major enzymes and
proteins involved with their functions are tabulated in Table 14.2.
Events in DNA Replication in E.coli
1.
Initiation
2.
Elongation
3.
Termination
DNA replication is depicted in Figure 14.14.
Initiation
·
DNA replication is initiated at replication origin known as oriC
(245 base pairs in E.coli).
·
Dna A protein molecules bind to the origin of replication.
·
Helicase (DnaB ) denatures the DNA helix by breaking the
hydrogen bonds between base pairs.
·
Many molecules of SSB (single stranded binding) proteins bind
cooperatively to single stranded DNA, stabilizing the separated strands and
preventing renaturation.
·
Gyrase (topoisomerase) releases the topological stress produced
by helicase.
·
RNA primers are synthesised by primase.
The separated polynucleotide strands are used as templates for
synthesis of complementary strands. The area of the DNA opened by helicase for
DNA synthesis is referred to as the replication fork. At the replication fork
there are four strands of DNA, two are conserved and two are newly synthesised.
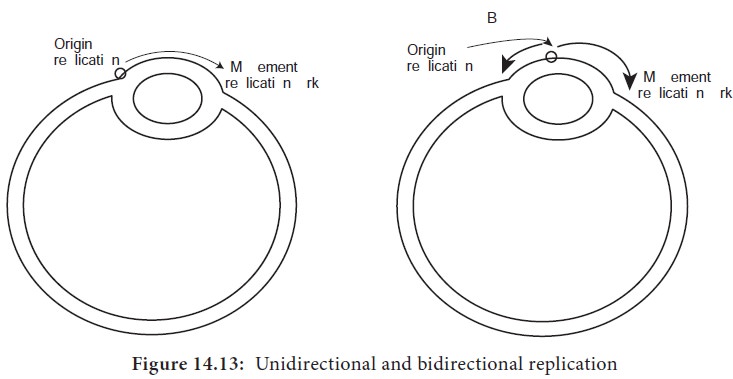
Replication may occur in either a unidirectional or bidirectional manner
(Figure 14.13) from each origin. Bidirectional replication can be explained as
two replication forks moving in opposite directions around the circular
chromosome. Both forks move along the double helix away from the origin of
replication in opposite directions and around the circular chromosome.
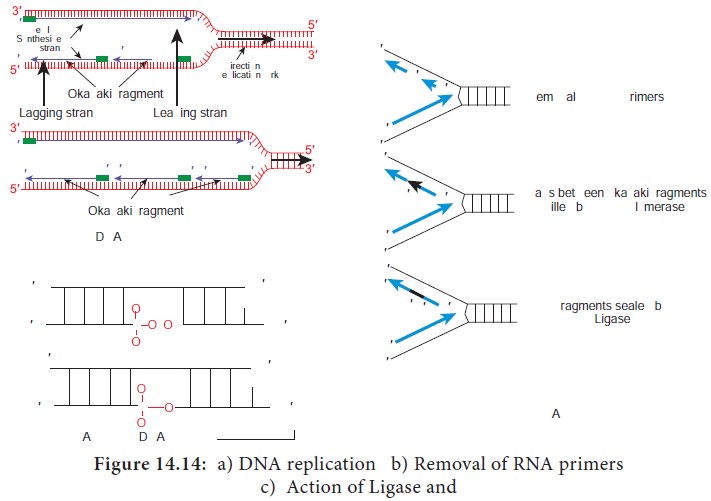
Elongation
·
DNA synthesis proceeds in a 5ʹ3ʹ direction( read as 5 prime to 3 prime).
· One strand is synthesised continuously and is known as leading strand.
·
Other strand is synthesised discontinuously and is known as
lagging strand.
·
The enzymes that are able to synthesise new DNA strands on a
template strand are called DNA polymerases. Kornberg was awarded Nobel Prize
for discovering DNA polymerase in 1956.
·
There are three known enzymes in Escherichia.coli viz., DNA polymerase I, II and III.
·
All of the known DNA polymerases can extend a
deoxyribonucleotide chain from a free 3ʹOH end, but none can initiate
synthesis.
·
Synthesis of DNA requires nucleoside triphosphates or
nucleotides - deoxyadenosinetriphosphate (dATP), deoxythymidinetriphosphate
(dTTP), deoxycytidinetriphosphate (dCTP), deoxyguanosinetriphosphate (dGTP).
When a nucleoside triphosphate bonds to sugar in a growing DNA strand, it loses
two phosphates.
·
RNA primer synthesised during initiation is removed and replaced
with DNA by DNA polymerase I
·
Sealing of the nick by DNA ligase which catalyses the formation
of phosphodiester bond between a 3ʹ hydroxyl end of one DNA fragment and 5ʹ
phosphate at the end of another strand.
The elongation phase of replication includes two distinct but
related operations
·
Leading Strand Synthesis
·
Lagging Strand Synthesis
Leading strand synthesis begins with the synthesis of short (10 to 60 nucleotide long) RNA primer at the
replication origin. Deoxyribonucleotides are added to this primer by DNA
polymerase III. Leading strand synthesis then proceeds continuosly, keeping
pace with the unwinding of DNA at the replication fork. The continuous strand
or leading strand is one in which 5ʹ3ʹ synthesis proceeds in the same direction as replication fork
movement.
Lagging strand or discontinuous strand is one in which 5'→3' DNA synthesis proceeds in the direction opposite to the direction of fork movement. This strand is synthesised as short fragments, known as Okazaki fragments named after R. Okazaki. Okazaki fragments range in length from a few hundred to a few thousand nucleotides depending on the cell types. Each okazaki fragment must be initiated by the action of primase. Once an Okazaki fragment has been completed its RNA primer is removed and replaced with DNA by DNA polymerase I and the nick is sealed by DNA ligase.
The fidelity of DNA replication is maintained by (1) base selection by the polymerase, (2) a 3'→5' proofreading exonuclease activity that is part of most DNA polymerases, and (3) specific repair systems for mismatches left behind after replication.
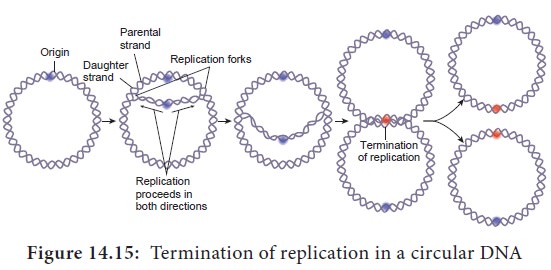
Termination
Eventually, the two replication forks of the circular E.coli chromosome meet at a terminus
region called Ter (for terminus). The Ter sequence function as binding sites
for protein Tus (Termination utilization substance). The Tus-Ter complex can arrest
a replication fork from only one direction. When either replication fork
encounters a functional Tus-Ter complex, it halts. The other fork halts when it
meets the first (arrested) fork (Figure 14.15).
Eukaryotic DNA Replication
Replication in Eukaryotic cells is more complex. The DNA molecules in Eukaryotes are considerably larger than those in bacteria and are organized into complex nucleoprotein structures (chromatin). The essential features of DNA replication are same in eukaryotes and prokaryotes. However, some interesting variations do occur. Initiation of replication in all eukaryotes requires a multisubunit protein. Multiple origins of replication are probably a universal feature in eukaryotic cells.
Like bacteria, eukaryotes have several types of DNA polymerases (Example: DNA
polymerase α[alpha] and DNA polymerase δ [delta]). Some have been linked to
particular functions, such as the replication of mitochondrial DNA. The
termination of replication on linear eukaryotic chromosomes involves the
synthesis of special structures called telomeres at the ends of chromosome.
Related Topics