Chapter: Medical Microbiology: An Introduction to Infectious Diseases: Host-Parasite Relationships
Corollaries of Microbial Pathogenicity
COROLLARIES OF MICROBIAL PATHOGENICITY
As noted, all parasitic microorganisms need to enter a host, find a unique niche, overcome local defenses, replicate, and be transmitted to a new host. Other factors have become ap-parent because of these pathogenic attributes. Some are more applicable to bacteria and fungi than to viruses and the larger parasites. The general principles are likely to be true for all pathogens.
1. Pathogenic microorganisms adapt to changes in the host’s biological and social behavior. Imagine the profound changes in the host – parasite relationship that must have occurred when humanoids began to live in communities and began to husband animals. The older diseases such as tuberculosis remained, but the increase in popula- tion density meant that “new” epidemic diseases could evolve. In recent times, we have seen new diseases emerge. Diseases such as TSS, Legionnaires’ disease, and nosocomial infections are a reflection, in part, of human progress. We need to remind ourselves that we live in a balanced relationship with microorganisms on this planet.Microorganisms will take advantage of any selective benefit that is made available to them to replicate and to establish themselves in a new niche. The advent of the birth control pill and the replacement of barrier contraception led to an enormous increase in sexually transmitted diseases. As humans increasingly impinge on other forms of life that have been largely isolated from human populations, there has been an in crease in “new” infectious diseases such as Lyme disease, and quite probably, AIDS As we have become more efficient at food production and mass global distribution there has been an increase, rather than a decrease, in food-borne infection and disease. One need no longer go to an esoteric place in the world to acquire traveler’s diarrhea,it can be readily acquired on imported food now at the corner food market!
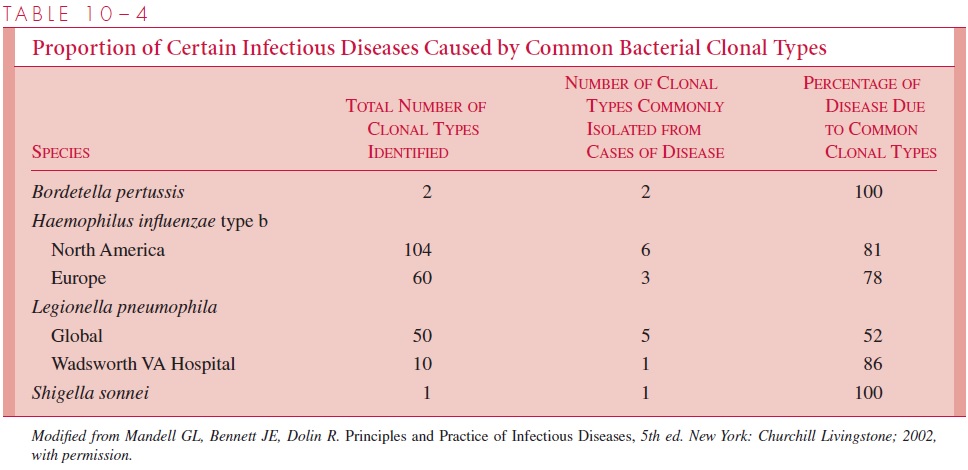
2. Pathogens are clonal.
Bacteria are haploid, as are viruses and some fungi. Conse- quently, there cannot be a helter-skelter amalgam of genes brought about by promis- cuous genetic exchange. If this were so, there would be no bacterial specialization and all would possess a consensus chromosomal sequence.
Thus, most bacteria (and viruses) have some degree of built-in reproductive isolation, except for members of their own or very closely related species (members of the same gene pool). In this way, diversity within the species through mutation can be maximized (usually by transformation or transduction), while conserving useful gene sequences. The end re-sult of husbanding of important genes during evolution is that at any given time in the world, many bacterial and viral pathogens are representatives of a single or, more of-ten, a relatively few clonal types that have become widespread for the (evolutionary) moment. Thus, all the strains of the typhoid bacillus that have been studied since humans learned to culture them belong to two basic clonal types (Table 10 – 4). When microbes establish a unique niche, they protect their selective advantage.
However, the bacterial gene pool must be expanded. Indeed, how could microor-ganisms have become pathogensin the first place or adapt to new potential niches? Bacteria have remarkable ways of expanding their genetic diversity, but they do so in a way that is consistent with their haploid lifestyle. From this corollary follows the next.
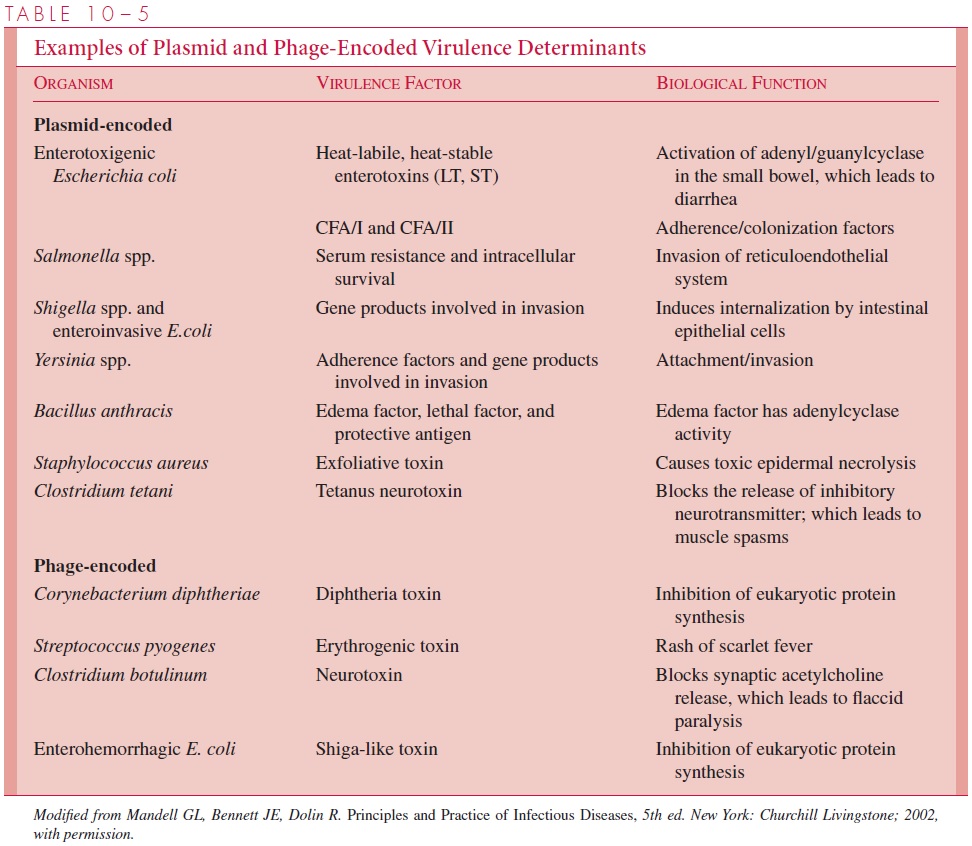
3. Pathogens often carry essential virulence determinants on mobile genetic elements.
It is now well established that many of the essential determinants of pathogenicity are actually replicated as part of an extrachromosomal element or as additions to the bacterial chromosome (Table 10 – 5). If haploid organisms must limit their genetic interactions to preserve their individuality, it is not surprising that new genes with im-portant new attributes are found on genetic elements that do not disrupt the organiza-tion of the bacterial chromosome. The interchange of plasmids and bacterial viruses among bacteria, coupled with transpositional (illegitimate) recombination between the extrachromosomal element and the chromosome, provides a means for microorgan-isms to exploit new genes in a haploid world. It is of some note that pathogenic deter-minants not found associated with a plasmid or a phage are often seen as duplicated genes or associated with transposon-like structures.
While it was clear for some time that mobile genetic elements played an essential role in the evolution of pathogenicity, only recently, with the advent of new DNA sequencing methods, have we learned that large blocks of genes found on the bacterial chromosome are associated with pathogenicity. These blocks of genes have been given the name patho-genicity islands (PAIs) to describe unique chromosomal regions found exclusively associ-ated with virulence. It is now generally believed that parts of a plasmid associated with virulence are likely PAIs as well. PAIs most often occupy large genomic areas of 10 to 200 or more kilobases. However, certain bacterial strains also carry insertions of smaller pieces of DNA with the attributes of PAIs, but they are only 1 to 10 kilobases in size and are referred to by some as pathogenicity islets. All of the available data are consistent
with the idea that horizontal gene transfer likely is mediated by phage or plasmids acquired
these large (and small) DNA sequences. However, the PAIs described thus far are not mobile in themselves. There is an eerie quality about the composition of many PAIs in the sense that they have a very different guanine _ cytosine content and codon usage as compared to the rest of the genome. The fact PAIs are so often associated with tRNA genes suggests that gene transfer from a foreign species is the likely origin. (In many prokaryotic and eukaryotic species, tRNA genes often act as the site of integration of foreign DNA.) Many PAIs have strikingly similar homologs in bacteria that are pathogenic for plants and animals and range from obligate intracellular parasites such as Chlamydia to free-living environmental opportunistic pathogens such as Pseudomonas aeruginosa.
In a bacterial genus that contains both pathogenic and nonpathogenic species, the attributes of pathogenicity are encoded on sequences that do not have any counterpart in the nonpathogen. It seems unlikely that pathogenicity arose as a result of long adaptation of an initially nonpathogenic organism to a more parasitic, host-dependent lifestyle. It is more likely that organisms inherited new gene sequences, often in a large block, that provided them with the capacity to establish themselves more efficiently in a host or to exploit some new niche within the host.
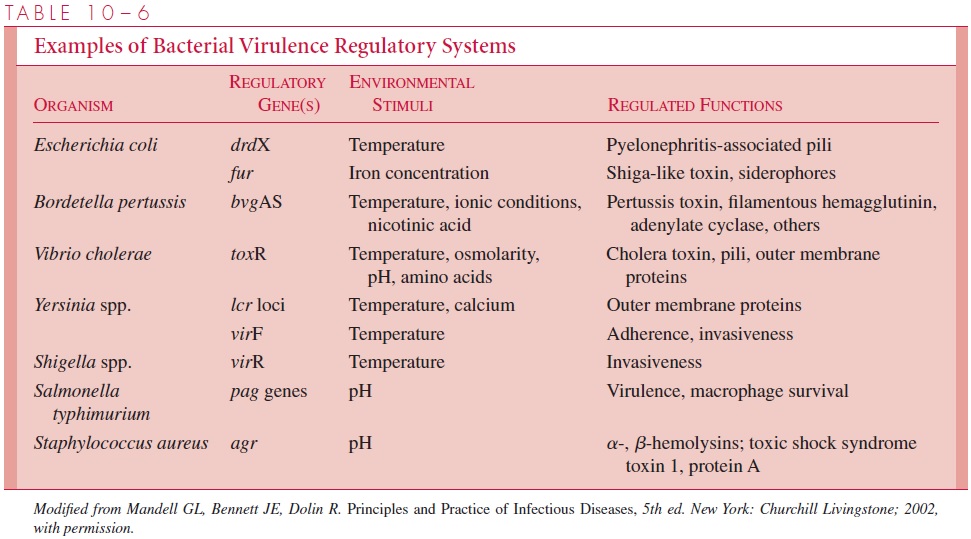
Bacteria, fungi, and larger parasites have evolved signaltransduction networks using environmental clues such as temperature, iron concentra- tion, and calcium flux to turn on genes important for pathogenicity (Table 10 – 6). It was puzzling to consider how a microorganism that makes potent toxins in the labora-tory could possibly spare any host it infected. It became clearer when we learned that toxin biosynthesis by the microbe is tightly regulated together with other genes to be activated only in particular sets of circumstances. Only in selective circumstances are genes involved in pathogenicity used and then often sparingly. The organism’s reac-tion to the host need only be sufficient to establish itself and replicate.
Related Topics