Chapter: Microbiology and Immunology: Antimicrobial Agents Therapy and Resistance
Mechanisms of Antibiotic Resistance - Resistance to Antimicrobial Drugs
Resistance to Antimicrobial Drugs
Bacterial resistance to drugs is a condition in which the bacte-ria
that were earlier susceptible to antibiotics develop resistance against the
same antibiotics and are not susceptible to the action of these antibiotics.
Antibiotic resistance among bacte-ria is a major concern in the treatment of a
patient. Emergence of antibiotic resistance to the old as well as new
antibiotics by bacteria is posing a major challenge in the treatment of
infections caused by bacteria.
Antibiotic resistance is seen more commonly in hospital-acquired
infections than in community-acquired infections. The antibiotic-resistant bacteria
are more commonly seen in hospital environment due to widespread use of
antibiotics in hospitals that select for these bacteria. These hospital strains
of bacteria are characterized by developing resistance to multiple antibiotics
at the same time. Common examples of such strains of bacteria showing drug
resistance include hospital strains of S.
aureus and Gram-negative enteric bacteria, such as Escherichia coli and
Pseudomonas aeruginosa. Resistance to multiple antibiot-ics is mediated by
plasmid-carrying several genes that encode enzymes responsible for the
resistance.
Mechanisms of Antibiotic
Resistance
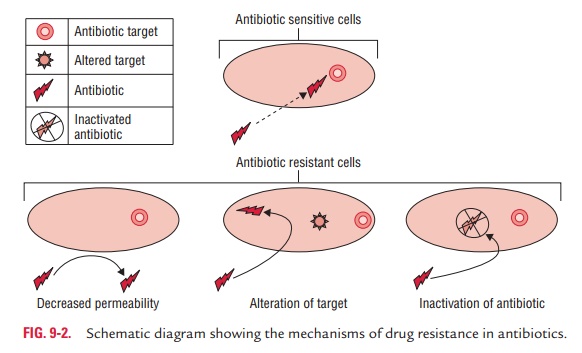
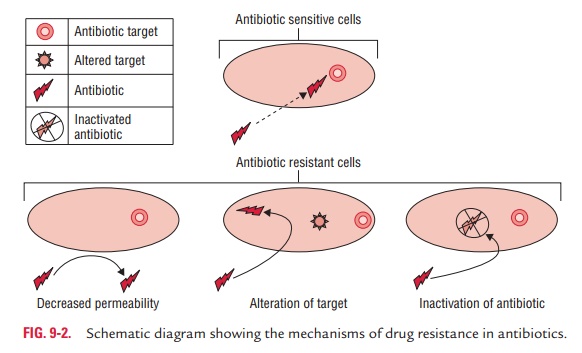
There are many different mechanisms by which
microorganisms might exhibit resistance to drugs (Fig. 9-2). These are (a) produc-tion of enzymes, (b) production of altered enzymes, (c) synthesis of modified targets, (d) alteration of permeability of cell
wall, (e) alteration of metabolic
pathways, and (f) efflux pump as
follows:
◗
Production of enzymes
Bacteria produce enzymes that inactivate antibiotics. For example,
penicillin-resistant staphylococci produce an enzyme -lactamase that destroys
the penicillins and cephalosporins by splitting the -lactam ring of the drug.
Gram-negative bacteria resistant to aminoglycosides, mediated by a plasmid,
produce adenylating, phosphorylating, or acetylating enzymes that destroy the
drug.
◗
Production of altered enzymes
Certain microorganisms develop an altered enzyme that can still
perform its metabolic function, but is much less affected by the drug. For
example, in trimethoprim-resistant bacteria, the dihydrofolic acid reductase is
inhibited far less efficiently than in trimethoprim-susceptible bacteria.
◗
Synthesis of modified targets
Certain bacteria produce modified targets against which the antibiotic has no effect. For example, a methylated 23S ribo-somal RNA can result in resistance to erythromycin, and a mutant protein in the 50S ribosomal subunits can result in resistance to streptomycin. Penicillin resistance in S. pneumoniae and enterococci is caused by the loss or alteration of PBPs.
◗
Alteration of permeability of cell wall
Some bacteria develop resistance to antibiotic by changing their
permeability to the drug in such a way that an effective intra-cellular
concentration of the antibiotic is not achieved inside the bacterial cell. For
example, P. aeruginosa develops
resistance against tetracyclines by changing its porins that can reduce the
amount of tetracycline entering the bacteria, thereby develop-ing resistance to
the antibiotics.
Resistance to polymyxins is also associated with a change in
permeability to the drugs. Streptococci have a natural perme-ability barrier to
aminoglycosides. This can be partly overcome by the simultaneous presence of a
cell wall–active drug, e.g., penicillin.
◗
Alteration of metabolic pathways
Bacteria may develop resistance by altering metabolic pathway that bypasses the reaction inhibited by the drug. For example, certain sulfonamide-resistant bacteria do not require extracel-lular PABA but, like mammalian cells, can utilize preformed folic acid.
◗
Efflux pumps
Efflux pumps have been found to be responsible for conferring
resistance to many groups of antibiotics including aminogly-cosides,
quinolones, etc. The major family of bacterial efflux pumps include ABC
(ATP-binding cassette) multidrug efflux pump, multidrug resistance and toxic
compound extrusion (MATE) efflux pumps, major facilitator superfamily efflux
(MFSE) pumps, etc.
Related Topics