Chapter: Plant Biology : Physiology and regulation
Methods in experimental plant science - Physiology and regulation
METHODS IN EXPERIMENTAL PLANT SCIENCE
Key Notes
Approaches to study plant function
Plant function and regulation was first studied by a combination of surgical, chemical and genetic approaches. Recently, research has made dramatic progress as a result of the techniques of molecular biology and the study of mutants, plants in which genetic change can be linked to change in form or function.
Mutants to study form and function
Mutants are generated by chemical treatment, radiation or inserting DNA into the genome (insertional mutagenesis). Considerable information on function and development can be gained by studying the phenotype of mutants and characterizing the mutated gene. Mutants can be used in almost all areas of experimental plant science, including development, photosynthesis and metabolism, and nutrition and water transport.
Molecular techniques
Molecular techniques provide information on the regulation and function of genes and their products. It is possible to determine and analyze the entire DNA sequence of genes, describe how they are regulated, observe the effects of both under- and over-expression and observe the effects of deliberately mutating the gene. Reporter gene constructs (in which a gene for a detectable product is fused with the gene of interest) have been of great value in studying function and regulation in cells and tissues.
Map-based cloning
The location of a gene (e.g. a developmental gene which has been mutated) on a chromosome can be identified using the technique of mapping. The crossover frequency of the gene is compared with genetic markers of known position on the chromosome.
Transposon tagging
Transposons are mobile pieces of DNA that insert into the genome, disrupting the gene into which they insert. When a gene disruption is found, it can be identified and characterized by looking for the transposon DNA and then characterizing the surrounding region.
Mutagenesis by T-DNA insertion
A transferred DNA (T-DNA) containing a selectable marker (a gene conferring a property that permits it to be readily detected) is inserted randomly into the genome by Agrobacterium-mediated transformation. When it inserts into a gene of developmental importance, that gene is disrupted. The selectable marker makes identification and
characterization of that gene straightforward.
Identifying a cloned gene
The product of mutagenesis experiments is usually an identifiable section of DNA containing the gene of interest, with markers for its identification. The DNA is then isolated either from a genomic library or from a cDNA library. It can then be sequenced and the complete amino acid sequence of the protein product determined. Comparison with data
in computer databases permits predictions about the nature and function of the protein to be made.
Analyzing patterns of gene expression
Once a gene has been identified, it is important to know where and when it is functional in the whole plant. In Northern blotting, the presence of the gene is identified by probing messenger RNA (mRNA) from the different parts of the plant or at different developmental stages. In in situhybridization, sections of tissue are probed. Reporter gene constructs provide a visible signal for the gene of interest.
Approaches to study plant function
Early studies of plant physiology and development involved growing plants in various environments, applying stimuli or carrying out surgery and observing changes in form and function. These were followed by biochemical analyses and the identification of compounds active in development. Similar methods were used to study photosynthesis, metabolism and transport, together with the use of radioactive tracer compounds. Studies like these, together with classical genetics, generated information on which much of our current understanding of the internal function of plants is built. However, a complete understanding of any plant system requires a knowledge of both the cellular components that carry out the processes observed and of the genes which encode those components. Understanding the genes includes understanding how they are regulated and how they function. Recently, plant biologists have combined the study of mutants with the techniques of molecular biology to gain detailed insights into the functioning of genes and gene products in development, photosynthesis, cell function, nutrient transport, metabolism and many other plant processes. In addition, by altering genes, it has been possible to describe the function of previously unknown cellular components. Model plants, and especially Arabidopsis(Topic E1) have been central to this work.
Mutants to study form and function
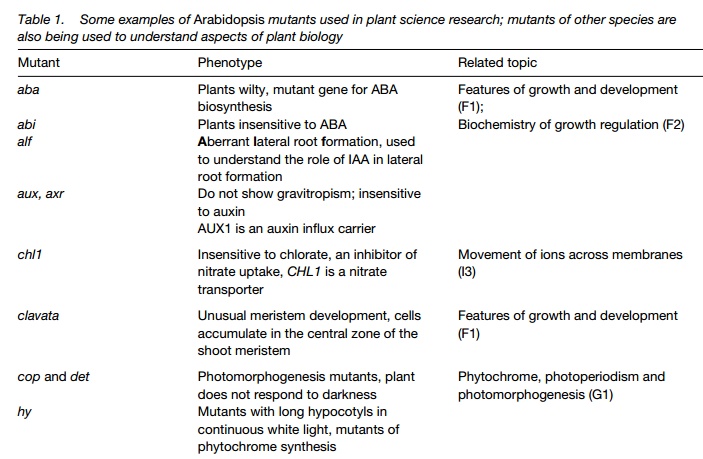
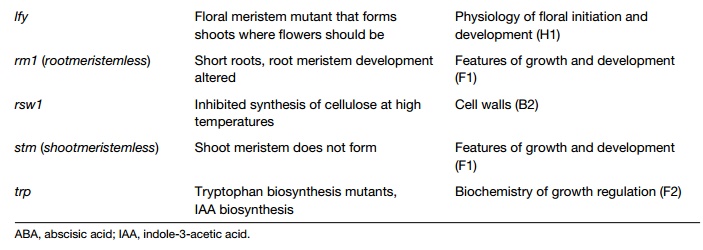
Mutants can be natural or generated in the laboratory by chemical treatment, commonly with ethylmethanesulfonate (ems), by irradiation of seedswith Xrays or by inserting DNA into the genome (insertional mutagenesis). Mutant seeds produced are known as M1 seeds. These are germinated, allowed to flower and self pollinate producing the M2 generation. By the rules of Mendelian genetics, one eighth of these plants will have a mutant phenotype characteristic of the mutagenized gene. They are carefully screened, which may involve scanning stems, leaves or flowers for unusual shape or color, determining the ability of the mutant to grow under specific conditions or more complex analysis. Many possibilities follow when plants showing interesting mutant phenotypes have been selected. Crosses are carried out to study the chromosomal location of the mutation; M2 seeds are germinated to establish that the mutant progeny run true to parental type. Most importantly, the mutated gene is analyzed, giving information on the gene, its regulation and the protein it encodes. Table 1 lists some of the many mutants that have been important in plant biology.
Molecular techniques
To understand a particular process within a plant, it is important to know the components involved and the ways that they are regulated. Regulation may involve altering gene expression (transcriptional regulation) or alteration of the
activity of constituents after gene expression (translational- and post-translational- modification). The activity of genes is altered when specific regulatorregions of the gene (promoters, repressors) are activated or inactivated. Theactivity of gene products is altered by a variety of regulated processes, whichalter the activity of cellular constituents. By analyzing many mutants, eachcomponent of such complex developmental systems can be described and therole of each of the contributors to development determined. In this process,identifying a gene is only the start. The gene must then be sequenced and bothits coding region (encoding the gene product) and non-coding regions (the regulatory regions) analyzed and compared with the wild-type gene. Sitedirected mutagenesis is used to deliberately alter these regions and the effects observed, confirming the role of the gene.
Map-based cloning
A genetic map describes the physical location of genes along a chromosome. The closer two genes are together, the less likely they are to separate and cross over in meiosis. When a mutant phenotype has been obtained, it is possible to map its location on a gene by measuring its frequency of recombination with respect to known genetic markers. Once the nearest marker has been found, techniques based on the mapping of chromosomes may be used to narrow down the location of the gene.
Transposon tagging
Transposable elements are mobile pieces of DNA present in eukaryotic genomes. They insert randomly into the genome and disrupt or inactivate the single gene into which they insert. Because the process is random, every mutant they produce is different, so they can be used to investigate almost any process. They are useful experimentally because they permit the experimenter to link genes and their effects. A plant produced has a phenotype altered by the disruption of a single gene, which can be recognized because it has a recognizable
piece of DNA – the transposable element – inserted into it. Having identified a plant showing a phenotype of interest, a genomic library of the transposon tagged stock is screened for the transposable element. Any clone selected by the screen contains the transposable element, with the mutated gene adjacent to it. This information is then used to screen a genomic library from a normal plant to give a full copy of the original gene.
Mutagenesis by T-DNA insertion
DNA transferred into a genome is termed T-DNA. It may be introduced by genetic engineering into the genome using an Agrobacterium tumifaciensbased vector (Topic O3). This T-DNA may be modified to include a selectable marker gene, e.g. for antibiotic resistance. This means that at a later stage, plants containing the T-DNA can be selected by growing them on agar containing antibiotic, as only the plants with the antibiotic resistance gene will be able to live. As with transposon tagging, when the T-DNA inserts into a gene of developmental importance, it disrupts its function and a mutant phenotype is observed. Identification of the gene is made easier because of the presence of the selectable marker. Stocks of T-DNA-tagged arabidopsis lines are now readily available for research.
Identifying a cloned gene
Once a gene of importance has been identified it can be cloned and sequenced. The product of a mutagenesis experiment is usually an identifiable section of DNA containing the gene of interest, together with appropriate markers for its identification. The DNA is then isolated. This may be from agenomic library, prepared by introducing random fragments of genomic DNA into a vector followed by propagation in either a yeast or a bacterial host. Alternatively, it
may be from a cDNA library, prepared by copying messenger RNA (mRNA) extracted from the plant to DNA using the enzyme reverse transcriptase. Once a cDNA encoding the gene or a genomic clone encoding the entire gene and its non-coding regions has been obtained, it can be analyzed further, by determining its DNA sequence. The sequence can be analyzed by comparison with other known sequences from computer databases, and the complete amino acid sequence of the protein product determined. The sequence can be manipulated and plants transformed to over- or under-express it or to express a mutant version, in order to analyze function further.
Analyzing patterns of gene expression
Once a gene has been identified, it is important to know where and when it is functional in the whole plant. Several techniques are available to do this. In Northern blotting, a specific probe is constructed using the gene. mRNA is
extracted from the different parts of the plant or at different developmental stages and separated on an agarose gel. The gel is then probed and levels of expression of the gene are determined. A similar, but more sophisticated technique, in situ hybridization, uses a similar probing technique with sections of tissue examined under the microscope. Reporter gene constructs are prepared by splicing a gene providing a visible signal to the gene of interest and transforming plants to express the construct. The reporter indicates both where and when the gene is active. Common reporters are:
●βglucuronidase (GUS), an enzyme which generates a colored product when the tissue expressing it is soaked in a substrate;
●Green fluorescent protein (GFP) a protein from the jelly fish Aequoria victoria which fluoresces visibly in living cells stimulated by light of appropriatewavelength. Recently, yellow (YFP) and cyan (CFP) fluorescent proteins have
been generated and can be used simultaneously to observe several gene products.
Reporter gene constructs may also be used to report on the level of biologically active molecules in a tissue. For instance, the GUS gene has been fused to a promoter region known to respond to auxin concentrations to create transgenic plants in which the concentration of auxin can be measured.
Related Topics