Chapter: Microbiology and Immunology: Laboratory Identification of Bacteria and Taxonomy
Identification of Bacteria
Identification of Bacteria
The complete identification of bacteria involves the following
steps: (a) morphology of bacterial
colony on solid medium, (b) growth in
liquid medium, (c) biochemical
reactions, (d ) antigenic structures,
(e) animal pathogenicity, ( f ) antibiotic sensitivity, ( g ) typing of bacterial strains, (h) rapid identification methods, and (i ) molecular methods.
Morphology of Bacterial Colony on Solid Medium
Morphology of the bacterial colony on solid medium depends on a
number of factors, such as nature of culture medium, tem-perature and time of
incubation, age of culture, and number of subcultures done. The characteristics
noted are shape, size, surface, edge, elevation, opacity, color, and hemolysis
of the colonies, as follows:
·
The colonies may be a few millimeters in size: pinhead size (Staphylococcus aureus) or pinpoint
(streptococci).
·
The shape may be circular or irregular, and surface of the colonies
may be smooth, rough, or granular.
·
The colonies on the medium may be flat, raised, low convex, convex,
or umbonate.
·
The edge may be entire, lobate, crenated, undulate, or ciliate.
·
The colonies may be transparent, translucent, or opaque.
·
Certain bacterial colonies are associated with production of
pigments and hemolysis around them.
Growth in Liquid Medium
Nutrient broth and peptone water are frequently used as liquid
media for growth of bacteria. The characteristics of bacterial growth in liquid
media provide some clue in presumptive iden-tification. For example,
streptococci produce granular deposits at the bottom of the liquid medium,
whereas most of the Gram-negative bacteria produce uniform turbidity. Pseudomonas spp. and other aerobic
bacteria tend to produce surface pellicles in liquid media. The pigment
production of some bacteria, such as Pseudomonas
aeruginosa can be appreciated in liquid media.
Smears prepared on glass slides from bacterial colonies grown on
either solid or liquid media are examined for bacte-ria by appropriate
staining. Gram staining is most widely used to differentiate between
Gram-positive and Gram-negative bacteria. Ziehl–Neelsen stain differentiates
acid-fast bacilli (e.g., Mycobacterium
tuberculosis, Mycobacterium leprae,
etc.) from nonacid-fast bacilli (Fig. 6-1). Albert’s stain is used to demon-strate
metachromatic granules in Corynebacterium
diphtheriae.
Biochemical Reactions
Biochemical reactions are very important in the identification of
bacterial isolates and in the identification of different bacterial species.
These tests depend on the presence of certain enzymes, such as catalase,
oxidase, urease, gelatinase, etc., produced by the bacteria. Commonly used
biochemical tests are described below:
◗ Catalase test
Catalase test is used to detect the presence of enzyme catalase in
a bacterium. The enzyme catalase catalyzes the breakdown of hydrogen peroxide
with the release of free oxygen. It is found in most aerobic and facultative
anaerobic bacteria. The main exception is Streptococcus
spp. It is not found in anaerobes.
Red blood cells contain catalase and their presence, therefore,
gives a false positive result.
Catalase test is primarily used to differentiate Staphylococcus and Streptococcus. In this test, a small amount of culture to be tested
is picked up from a nutrient agar plate with a sterile platinum loop or glass
rod and this is inserted into hydrogen peroxide solution (3%) held on a slide
or in a tube. Immediate production of air bubbles in the solution denotes a
positive test and no bubbles indicate a negative test.
◗ Oxidase test
This test determines the presence of enzyme oxidase in many
bacteria. The enzyme oxidase catalyzes the oxidation of reduced cytochrome by
molecular oxygen. Kovac’s oxidase reagent that contains tetramethyl-p-phenylenediamine dihydrochloride is
the main reagent used in the oxidase test. The dye serves as an alternate
substrate for the cytochrome oxidase reaction. In the reduced state, the
reagent is colorless but when oxidized, it becomes purple. Oxidase test can be
performed by several methods that include:
1.
Dry filter paper method,
2.
Wet filter paper method, and
3.
Plate method.
The dry filter paper method is performed by impregnating strips of
filter paper with 1% Kovac’s oxidase reagent. The paper is smeared with the
bacterial colonies to be tested by a glass rod. In a positive test, the smeared
area on the filter paper turns deep purple within 10 seconds. No color change
indicates nega-tive test (Color Photo 4).
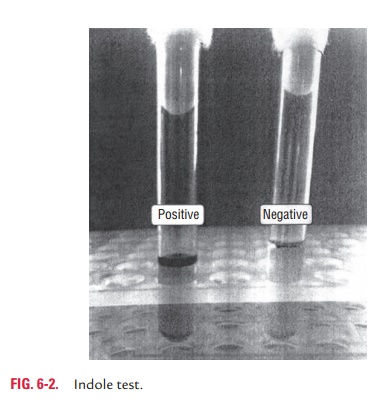
◗ Indole test
Indole test is used to detect the ability of bacteria to decompose
amino acid tryptophan to indole, which accumulates in the medium. Tryptophan or
peptone broth is the medium used for indole test (Color Photo 5). The test is
performed by inoculating the medium with bacteria, incubating at 37°C for 24–48
hours. Then, 5 drops of Kovac’s reagent containning amyl or isoamyl alcohol, p-dimethyl amino benzaldehyde, and
concentrated hydro-chloric acid are added to the inoculated medium. Positive
test is indicated by formation of a red ring at the surface of the medium. No
color change indicates a negative test (Fig. 6-2).
Precautions in interpretation of indole test are as follows: (a)
cultures to be tested for indole production need to be incubated aerobically
and (b) the optimum pH for tryptophanase activity is alkaline (pH 7.4–7.8),
hence a decrease in pH results in decreased indole production and gives a
possible false negative reaction.
◗ Carbohydrate fermentation test
Carbohydrate fermentation test is done to determine the ability of
a bacterium to ferment a specific carbohydrate incorporated in a basal medium,
producing acid or acid with visible gas. The carbohydrates include glucose, lactose,
sucrose, maltose, etc. The sugar medium contains 1% sugar. Andrade’s indicator
is a solution of acid fuchsin to which sodium hydroxide is added. It is a pH
indicator that is added to the basal medium, which indicates the formation of
organic acids.
The test is performed by inoculating the sugar media with bacteria,
incubating at 37°C for 18–24 hours. The change of the color to pinkish red
(acidic) is considered as a positive test result, whereas yellow to colorless
(alkaline) sugar indicates negative test result. Production of gas is indicated
by appear-ance of gas bubbles in Durham’s tube.
◗ Oxidation–fermentation test
Oxidation–fermentation test (OF test) determines the oxida-tive or
fermentative metabolism of a carbohydrate by a bacte-rium. The OF test is used
to determine whether a bacterium has the enzymes necessary for the aerobic
breakdown of glucose (i.e., oxidation) and/or for the fermentation of glucose.
The test differentiates Enterobacteriaceae (fermenters) from the mem-bers of
the family Pseudomonadaceae (the nonfermenters).
◗ Kligler’s iron agar/triple sugar iron agar test
Kligler’s iron agar (KIA) and triple sugar iron agar (TSI) tests
are used to determine the ability of an organism to attack a specific
carbohydrate incorporated into a basal growth medium, with or without the
production of gas, along with production of hydrogen sulfide. KIA medium
contains two carbohydrates: lactose and glucose in a ratio of 10:1. TSI
con-tains a third carbohydrate, i.e., sucrose, in addition to lactose and
glucose.
The test is performed by inoculating KIA or TSI with an inoculating
needle by stabbing the butt and streaking the slant and then incubating at 37°C
for 18–24 hours. During incubation, the bacterium first utilizes glucose and
then lactose and sucrose. After 18–24 hours, the glucose concentration is
depleted in the slant and in the butt. The bacteria start oxidative degradation
of the peptone present in the slant, resulting in the production of alkaline
by-products, thereby changing the indicator to a red color. Anaerobic fermentation
of glucose in the butt produces a large volume of acid, which neutralizes the
alkalinity caused by peptone degradation; hence the butt continues to remain
yellow. Yellow color (acidic), therefore, indicates fermentation of
carbohydrates and red color (alkaline) indicates no fermentation. Presence of
bubble in the butt indicates production of gas during fermentation of
carbohydrates.
Certain bacteria produce H2S, which is detected as black
precipitate that blackens the slant and butt of the medium. The medium is
turned black due to combination of H2S with ferric ions, from ferric
salts present in the medium, to form ferrous sulfide as black precipitates.
Three basic fermentation patterns are observed on KIA medium after
incubation at 37°C for 24 hours:
1.
Fermentation of glucose only
(alkali/acid or K/A reaction; K denotes alkaline reaction and A denotes acidic
reaction): After 18–24 hours, the low glucoseconcentration is completely used
up, and the organism starts to utilize the peptones present in the medium,
resulting in an alkaline pH in slant (red). In the butt, a yellow color exists
due to anaerobic degradation of glucose.
2.
Fermentation of lactose and
glucose (acid/acid or A/A reaction): Lactose is present in 10 times the amount
ofglucose. In 18–24 hours, the lactose is not depleted and therefore acidic
conditions exist in the butt and slant.
3.
Neither lactose nor glucose
fermented (alkali/alkali or K/K reaction): Many Gram-negative, nonenteric bacilliare
unable to ferment glucose or lactose. A reaction of K/K is a result of aerobic
catabolism of peptone by the organism.
H2S production and carbohydrate fermentation patterns
are generally characteristic for specific bacterial groups, especially the
Enterobacteriaceae.
◗ Urease test
Urease test is used to determine the ability of an organism to
split urea to ammonia by the enzyme urease. Production of ammonia makes the
medium alkaline; thus the indicator phe-nol red changes to red or pink in
color.
The test is performed in Christensen’s urease medium. The medium is inoculated with the bacterial colony and incubated at 37°C. Urease-positive bacteria produce a pink color.
◗ Citrate test
Citrate test is used to demonstrate the ability of an organism to
utilize citrate as the sole source of carbon for metabolism. Koser’s citrate
medium (a liquid medium) and Simmon's citrate medium (a solid medium) are used
for the test. In this test, either of the medium is inoculated with the
bacteria and then incu-bated at 37°C overnight. Growth on the Simmon's medium
is accompanied by a rise in pH to change the medium from its initial green
color to deep blue. Hence, growth with blue color on the slant indicates
positive test and no growth without any color change indicates negative test.
The medium needs to be lightly inoculated (from plate cultures, not from a
broth) to avoid a carryover of nutrients, which may lead to a false positive
result. In Koser’s liquid medium, turbidity indicates positive test and absence
of turbidity indicates negative test.
◗ Phenylalanine deaminase test
Phenylalanine deaminase test indicates the
ability of an organism to deaminate phenylalanine to phenylpyruvic acid (PPA),
which reacts with ferric salts to give green color. This test is also called
PPA test. The test is performed by inoculating the medium containing
phenylalanine by the bacteria. After overnight incubation at 37°C, a few drops
of 10% ferric chloride solution are added to the inoculated medium. If PPA is
produced, the medium becomes green in color due to combination of ferric
chloride with PPA, which indicates a positive test. Absence of any color change
indicates a negative test. PPA-positive bacteria are Proteus spp., Providencia spp.,
and Morganella spp.
◗ Nitrate reduction test
Nitrate reduction test is used to determine the presence of enzyme
nitrate reductase in the bacteria. The enzyme reduces nitrate to nitrites or
free nitrogen gas. The test is carried out by inoculating the broth containing
1% potassium nitrate (KNO3) and incubating at 37°C up to 5 days.
Then 1–2 drops of a reagent that consists of a mixture of 1 mL of naphthylamine
and 1 mL of sulfanilic acid is added. Red color developing within a few minutes
signifies positive reaction, while absence of color indicates negative
reaction.
◗ Methyl red test
Methyl red (MR) test detects the ability of an organism to produce
and maintain stable acid end products during the fermentation of glucose,
thereby maintaining a sustained pH below 4.5. The test is performed by
inoculating a glucose phosphate broth and incubating it at 37°C for 2–5 days.
Five drops of 0.04% of MR solution are then added to the inocu-lated medium,
mixed, and the result is read immediately. Development of red color denotes a
positive test, and yellow color indicates negative test. E. coli, Yersinia spp.,
and Listeriamonocytogenes are the
examples of MR-test-positive bacteria.
It should be noted that if the MR test is performed too early, the
results may produce a false positive reaction. This is because MR-negative
organisms may not have adequate time to completely metabolize the initial acid
products that have been produced during the fermentation of glucose.
◗ Voges–Proskauer test
Voges–Proskauer
(VP) test detects the production of acetyl methyl carbinol (acetoin), a natural
product formed from pyruvic acid in the course of glucose fermentation. The
acetoin, in the presence of alkali and atmospheric oxygen, is oxidized to
diacetyl that reacts with alpha-naphthol to produce red color. This test is
performed by inoculating the glucose phosphate broth with the organism and
incubating at 37°C for 48 hours. Then approximately 3 drops of alpha-naphthol
is added followed by addition of 1 drop of 40% potassium hydroxide. The
reagents are mixed well and are allowed to stand for 30 minutes. In positive
test, pink color appears in 2–5 minutes, deepening to magenta in half an hour.
The solution remains colorless for 30 minutes in negative test.
Staphylococcus
spp., V. cholerae biotype eltor, Klebsiella spp., and Enterobacter spp. are the common
examples of VP-test-positive bacteria.
◗ Hydrogen sulfide production
These tests are carried out to demonstrate H2S produced
by certain bacteria. Production of H2S is demonstrated by
inoculating the bacteria in the media containing lead acetate, ferric ammonium
citrate, or ferrous acetate and incubating overnight at 37°C. H2S-producing
bacteria by their enzymatic actions produce H2S from
sulfur-containing amino acids in the medium. Production of sulfur produces
black color in the media, visible to the naked eye.
Filter paper strip is another method of demonstration of production
of H2S. In this method, filter paper strip impreg-nated with lead
acetate is kept between the cotton plug and the medium in the tube. Production
of H2S is indicated by blackening of the paper.
P. mirabilis, P. vulgaris, and Salmonella spp.
are some examplesof the H2S-producing bacteria.
Antigenic Structures
Agglutination of biochemically confirmed bacteria with specific
antisera facilitates further identification of the isolated bacteria.
Agglutination test is used in the identification of presumptive isolates of
pathogens (e.g., Salmonella) from
clinical samples. It is also used for the grouping of beta-hemolytic
streptococci.
Animal Pathogenicity
Some pathogenic bacteria and their bacterial metabolites produce
characteristic lesions in laboratory animals. The most commonly employed
animals are rats, guinea pigs, mice, and rabbits. These animals, depending on
the organisms to be tested, may be inoculated by subcutaneous, intramus cular,
intravenous, intraperitoneal, or intracerebral routes. The iden-tification of
the bacteria is made depending on the postmor-tem findings and cultural
properties of the bacteria. For example, guinea pigs are commonly used for
performing animal pathogenicity of C.
diphtheriae, Clostridium perfringens,
and M. tuberculosis.
Antibiotic Sensitivity
Determination of antibiotic sensitivity of an
isolate from a patient is essential for the choice of drug therapy. In some
cases, sensitivity of an organism to a particular agent helps in the
identification, e.g., Streptococcus
pyogenes is sensitive to bacitracin and Streptococcus
pneumoniae to optochin.
Typing of Bacterial Strains
The ability to discriminate between similar strains of bacte-ria
may be important in tracing sources or modes of spread of infection in a
community. Typing methods are widely used for epidemiological studies. These
include (a) phenotypic techniques and
(b) genotypic techniques.
◗ Phenotypic techniques
Phenotyping techniques depend on various observable properties of
bacteria, which are discussed as follows:
·
Biotyping: It relies on a set of
biochemical reactions to dis-tinguish different strains within a given species.
Antimicro-bial susceptibility testing is an example of this type.
·
Serotyping: Different strains of
organisms of the samespecies can be differentiated based on the difference in
the expression of antigenic determinants on the cell surface.
·
Bacteriocin
typing: This is used in case of bacterial species
forwhich a number of lytic bacteriophages have been identified.
·
Phage typing: This has been the mainstay of
strain discrim-ination and is widely used in epidemiological studies.
◗ Genotypic techniques
Genotypic techniques depend on differences related to the genome of
bacteria. Genotypic techniques employed to differentiate strains of bacteria
include plasmid profile analysis and restriction endonuclease analysis of
chromosomal DNA.
Rapid Identification Methods
Automated methods are now available, which take only hours for
characterization of isolates. These include detection of specific enzymes,
toxins, antigens, or metabolic end prod-ucts. Obligate anaerobes can be
identified by gas liquid chromatography of short-chain fatty acids produced by
them.
Latex particle agglutination, coagglutination, direct fluo-rescent
antibody test, and dot enzyme-linked immunosorbent assay (ELISA) are the most
frequently used techniques in the clinical laboratory for rapid detection of
microbial antigens directly in clinical specimens. Antibody to a specific
antigen is bound to latex particles or to a heat-killed and treated protein
A-rich strain of S. aureus to produce
agglutination.
Molecular Methods
Molecular methods, such as nucleic acid probes, polymerase chain
reaction (PCR), and other amplification procedures are also used increasingly nowadays
for identification of micro-organisms. Genetic probes are based on the
detection of unique nucleotide sequences with the DNA or RNA of a
microorganism. Hybridization of the sequence with a complementary sequence of
DNA or RNA follows cleavage of the double-stranded DNA of the microorganism in
the specimen. PCR has major applications in the detection of infections due to
microorganisms that are difficult to culture (e.g., the human immunodeficiency
virus) or that have not as yet been successfully cultured (e.g., the Whipple’s
disease bacillus).
Related Topics