Chapter: Basic Concept of Biotechnology : Plant Transgenics: Genetic Engineering Approch to Devlop Biotic Stress Resistance Plants
RNAi Technology
RNAi TECHNOLOGY
A decade has passed since the discovery that double-stranded RNA molecules (dsRNA) can trigger silencing of homologous genes, and it is now clear that RNA-mediated gene silencing is a widely conservedcellular mechanism in eukaryotic organisms. The definition says RNA induced silencing of specific miRNA / gene is called RNA interface also known as gene silencing. In plants it is called as Post transcriptional silencing and in fungi it is known Quelling. RNAi mechanism evolved to provide immunity against various viruses and transposons. RNA-mediated gene silencing can be categorized into two partially overlapping pathways; the RNA interference (RNAi) pathway and the micro-RNA (miRNA) pathway. RNAi is triggered by either endogenous or exogenous dsRNA, and silences endogenous genes carrying homologous sequences at both the transcriptional and posttranscriptional levels. In contrast, the miRNA pathway is triggered by mRNAs transcribed from a class of non-coding genes. These mRNAs form hairpin-like structures, creating double-stranded regions in a molecule (pre-miRNA). In either pathway, dsRNA molecules are processed by Dicer RNase III proteins into small RNAs, which are then loaded into silencing complexes. In the RNAi pathway, small RNAs are called short interfering RNAs (siRNAs) and are loaded into RNA induced silencing complexes (RISC) for post-transcriptional silencing, or RNA-induced initiation of transcriptional gene silencing (RITS) complexes for transcriptional silencing. In contrast, miRNAs (small RNAs in the miRNA pathway) are loaded into miRNA ribonucleoparticles (miRNPs) for a review of silencing complexes). dsRNA binding motif (dsRBM) proteins, such as R2D2 and Loquacious, help small RNAs to be loaded properly into silencing complexes. Using the small RNA as a guide, silencing complexes find target mRNAs and cleave them (in the case of RISC) or block their translation (in the case of miRNP). RITS is involved in transcriptional silencing by inducing histone modifications. Argonaute family proteins are the main components of silencing complexes, mediating target recognition and silencing. The RNAi pathway and miRNA pathway are essentially parallel, using related but distinct proteins at each step.
RNAi is sequence-specific gene silencing at the post-transcription level, induced by double stranded RNA (dsRNA). The mediators of sequence-specific mRNA degradation are 21–23 nucleotides long, small interfering RNAs (siRNA) generated from longer dsRNAs by ribonuclease III cleavage activity (Fire et al., 1998; Cogoni and Macino, 2000; Hannon, 2002; Agrawal et al., 2003). The presence of RNAi machinery in insects has already been confirmed in earlier studies by introducing long dsRNA molecules into insects by means of using insect cell line (Zhang and Shi, 2002) or through injection into insect body (Soreq et al., 1994) and also via forced feeding (Dong and Friedrich, 2005). That siRNAs are powerful agents for gene silencing, even at low concentrations, and mediate post-transcriptional degradation. Systemic RNAi was first described in plants as spread of post transcriptional gene silencing. The first animal in which RNAi was shown to work systemically was C. elegans, where it has been thoroughly investigated.
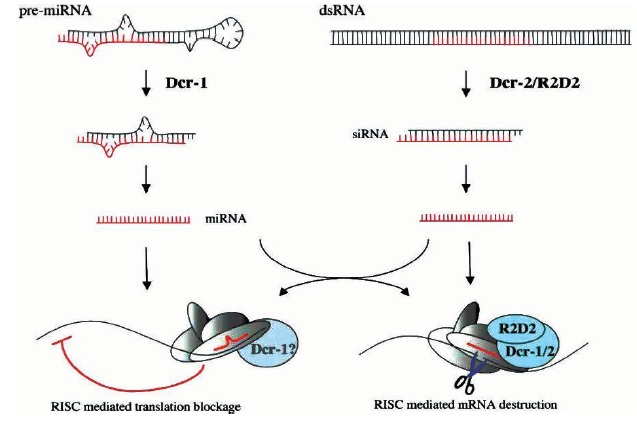
processed from double-stranded precursor molecules by Dcr-1 and Dcr-2, respectively, and stay attached to Dicer-containing complexes, which assemble into RISC. The degree of complementarity between the RNA silencing molecule (in red) and its cognate target determines the fate of the mRNA: blocked translation or immediate destruction.
Although current understanding of RNAi activity cannot provide us with a precise prediction of potency to each individual siRNA. Once the bioinformatics part is done, candidate siRNA can then be synthesized and tested in cell culture systems for knockdown efficiency. The off-target effect can also be checked with a microarray assay. The final goal of this stage is to identify several siRNAs that show high knockdown efficiency and minimal off-target effect at nanomolar or lower concentrations.
Systematic Application of RNAi Technology
A growing interest is to exploit RNAi technology for systematic biology and functional genomics research to knockdown gene expression in whole-genome or whole-pathway scales. In Conclusions RNAi technology is a very timely invention in the era of post genome to serve as one of the most powerful tools for reverse genetics, functional genomics, and systematic biology. Although we should keep in mind that continuous improvements and modifications are necessary to make RNAi technology more potent, it already revolutionizes our way of doing biology and medicine. The explosion of RNAi technology actually benefited from the previous development of antisense technology. The discovery of the RNAi/miRNA pathway opens the door to RNAi technology, and further characterization of this pathway really facilitates the development of RNAi technology step by step.
Related Topics