Chapter: Pharmaceutical Biotechnology: Fundamentals and Applications : Genomics, Other “Omics” Technologies, Personalized Medicine, and Additional Biotechnology Related Techniques
“Omics” Enabling Technology: Bioinformatics
“Omics” Enabling Technology: Bioinformatics
Structural genomics, functional genomics, proteomics, and other “omic”
technologies have generated an enormous volume of genetic and biochemical data
to store. The entire encoded human DNA sequence alone requires computer storage
of approximately 109 bits of information: the equivalent of a thousand 500-page books!
GenBank (managed by the National Center for Biotechnology Information, NCBI, of
the National Institutes of Health), the European Molecular Biology Laboratory
(EMBL), and the DNA Data Bank of Japan (DDBJ) are three of the many centers
worldwide that collaborate on collecting DNA sequences. These databanks, (both
public and private) store tens of millions of sequences (Martin et al., 2007).
Metabolic databases and other collections of biochemical and bioactivity data
add to the complexity and wealth of information (Olah and Oprea, 2007). Once
stored, analyzing the volumes of data, (i.e., comparing and relating information
from various sources) to identify useful and/or predictive characteristics or
trends, such as selecting a group of drug targets from all proteins in the
human body,presents a Herculean task. This approach has the potential of
changing the fundamental way in which basic science is conducted and valid
biological conclusions are reached (Baxevanis, 2001).
Scientists have applied advances in information technology, innovative
software algorithms and mas-sive parallel computing to the on-going research in
genetics, genomics, proteomics, and related areas to give birth to the fast
growing field of bioinformatics (Emmett, 2000; Felton, 2001; Watkins, 2001;
Lengauer and Hartman, 2007). Bioinformatics is the application of computer
technologies to the biological sciences with the object of discovering
knowledge. With bioinformatics, a researcher can now better exploit the
tremendous flood of genomic and proteomic data, more cost-effectively data
mining for a drug discovery “needle” in that massive data “haystack.” In this
case, data mining refers to the bioinformatics approach to “sifting” through
volumes of raw data, identifying and extracting relevant information, and
developing useful relationships among them. Modern drug discovery will utilize
bioinformatics techniques to gather information from multiple sources (such as
the HGP, functional genomic studies, proteomics, pheno-typing, patient medical
records, and bioassay results including toxicology studies), integrate the
data, apply life science developed algorithms, and generate useful target
identification and drug lead identifica-tion data. Another goal of
bioinformatics is to be able to study the molecules and processes discovered by
genomics and proteomics research in silico: that
is, to be able to predict chemical and physical structure and properties by
computer (Rashidi and Buehler, 2000).
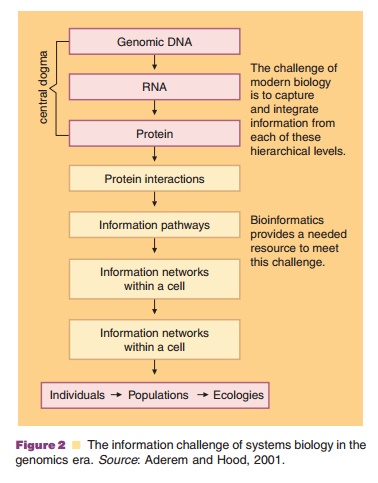
Bioinformatics in its multi-faceted implementa-tions may be thought of
as a technique of “electronic biology” (eBiology), conceptual biology, in silico biol-ogy, or computational biology (Blagosklonny and Pardee, 2002). A
data-driven tool, the integration of bioinformatics with functional knowledge
of the com-plex biological system under study remains the critical of any of
the omic technologies described above and to follow. As seen in Figure 2, the
hierarchy of information collection goes well beyond the biodata contained in
the genetic code that is transcribed and translated.
Related Topics