Chapter: Pharmaceutical Biotechnology: Fundamentals and Applications : Genomics, Other “Omics” Technologies, Personalized Medicine, and Additional Biotechnology Related Techniques
Proteomics, Structural Proteomics, and Functional Proteomics - “Omics” Technologies
Proteomics, Structural Proteomics, and Functional
Proteomics
Functional genomics research will provide an unprece-dented information
resource for the study of biochemical pathways at the molecular level.
Certainly a large number of the ~25,000
genes identified in sequencing the human genome will be shown to be
functionally important in various disease states (see druggable genome
discussion above). This will result in the identification of a vast array of
proteins implicated as playing pivotal roles in disease processes (Evans,
2003). These key identified proteins will serve as potential new sites for
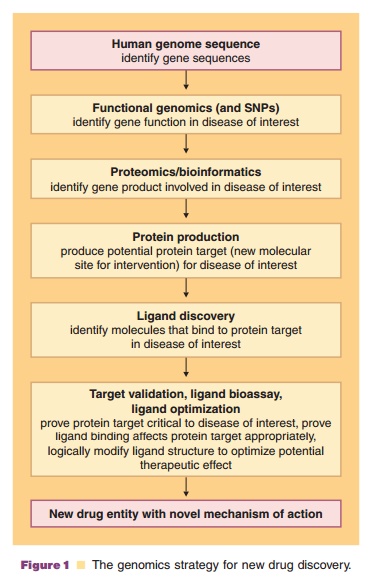
therapeutic intervention (Fig. 1). The research area called proteomics seeks to
define the function and correlate that with expression profiles of all proteins
encoded within an organism’s genome or “proteome” (Edwards et al., 2000;
Kreider, 2001; Voshol et al., 2007). The ~25,000
human genes can produce 100,000 proteins. The number, type and concentration
may vary depending on cell or tissue type, disease state, and other factors.
The proteins’ function(s) aredependent on the primary, secondary, and tertiary
structure of the protein and the molecules they interact with. Less than 20
years old, the concept of proteomics requires determination of the structural,
biochemical and physiological repertoire of all proteins. Proteomics is a
greater scientific challenge than genomics due to the intricacy of protein
expression and the complexity of 3-D protein structure (structural proteomics)
as it relates to biological activity (functional proteomics) (Saeks, 2001).
Protein expression, isolation, purification, identification, and
characterization are among the key procedures utilized in proteomics research.
To perform these procedures, technology platforms such as 2-D gel
electrophoresis, mass spectrometry, chip-based micro-arrays, X-ray
crystal-lography, protein nuclear magnetic resonance (NMR), and phage displays
are employed. Initiated in 2002, the Human Proteome Organization (HUPO)
recently completed the first large-scale study to characterize the human serum
and plasma proteins, i.e., the human serum and plasma proteome (States et al.,
2006). Pharmaceutical scientists anticipate that many of the proteins
identified by proteomic research will be entirely novel, possessing unknown
functions. This scenario offers not only a unique opportunity to identify
previously unknown molecular targets, but also to develop new ultrasensitive
diagnostics to address unmet clinical needs (Petricoin et al., 2002). Today’s methodology
does not allow us to identify valid drug targets and new diagnostic
methodologies simply by examining gene sequence information. However, “in silico proteomics”, the computer-based prediction of 3-D protein structure,
intermolecular interactions, and functionality is currently a very active area
of research (Roberts and Swinton, 2001; Voshol et al., 2007).
Often, multiple genes and their protein products are involved in a
single disease process. Since few proteins act alone, studying protein interactions
will be paramount to a full understanding of functionality. Also, many
abnormalities in cell function may result from over-expression of a gene and/or
protein, under-expression of a gene and/or protein, a gene mutation causing a
malformed protein, and post-translation modification changes that alter a
protein’s function. Therefore, the real value of human genome sequence data
will only be realized after every protein coded by the 25,000 genes has a
function assigned to it.
Related Topics