Chapter: Biotechnology Applying the Genetic Revolution: Forensic Molecular Biology
Using Repeated Sequences in Fingerprinting
USING
REPEATED SEQUENCES IN FINGERPRINTING
A variation of DNA fingerprinting is to
look at regions of the DNA that contain variable number tandem repeats (VNTRs). As discussed, this means that sequences of DNA are repeated multiple times
and that different people have different numbers ofmrepeats. Repeat sequences
vary greatly in length; however, for forensic purposes, relatively short
repeated sequences are now generally used and are known as short tandem repeats
(STRs) —see later discussion. VNTRs usually occur in noncoding regions of DNA.
Hence, using VNTRs protects privacy in the sense that an individual’s coding
DNA is not revealed during forensic investigations. VNTRs may be visualized by
using restriction enzymes tocut out the DNA segment containing the VNTR,
followed, as before, by separation of DNA bands by gel electrophoresis and
visualization by Southern blotting. Alternatively, the DNA fragments for VNTR
analysis may be generated by PCR (see later discussion). Figure 24.6 shows
corresponding DNA fragments from three individuals who differ in the number of repeats
in the same VNTR. Consequently the length of the fragment differs from person
to person.
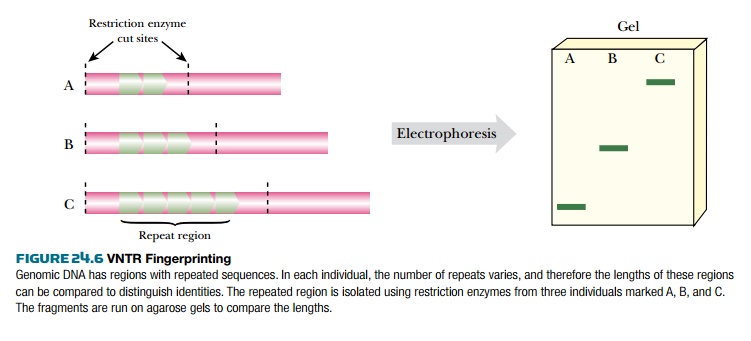
There is often an enormous variation
between people in the number of repeats at any particular VNTR site. So there
is a very low likelihood of two people matching exactly, or, if you prefer, a
high probability they will differ. Some VNTRs have 100 to 200 different variants,
making them very useful for forensic analysis. Although VNTRs are not genuine genes,
their variants are regarded as alleles and so VNTRs are considered to be
“multiallelic” systems.
One practical problem here is that
multiple tandem repeats give so many closely packed bands that standard agarose
gel electrophoresis cannot discriminate the different fragments. Different
types of gels such as polyacrylamide may resolve closely spaced bands.
Alternatively, the fragments can be separated on a gradient gel.
The original DNA fingerprints, invented
by Alec Jeffreys in England in 1985, used highly variable VNTRs with long
repeat sequences. DNA was isolated and cut with restriction enzymes because PCR
had not been invented. The cut fragments were probed using MLP to generate the
early DNA fingerprints.
The STR (short tandem repeat) is a
subcategory of VNTR in which the repeat is from two to six nucleotides long. Most
STRs are not as variable in the number of repeats as VNTRs with longer unit
sequences. Many STR sequences have only 10 to 20 alleles and hence cannot provide
unique identification alone. However, many STR loci are available, and if
several are analyzed simultaneously, this will provide enough data that the
pattern would be unique for each individual. Today, STR analysis is done using
PCR (see later discussion) to generate the DNA fragments.
Related Topics