Chapter: Genetics and Molecular Biology: Protein Structure
Structures within Proteins
Structures within Proteins
It is useful to focus attention on particular
aspects of protein structures. The primary structure of a protein is its linear
sequence of amino acids. The local spatial structure of small numbers of amino
acids, inde-pendent of the orientations of their side groups, generates a
secondary structure. The alpha helix, beta sheet, and beta turn are all
secondary structures that have been found in proteins. Both the arrangement of
the secondary structure elements and the spatial arrangement of all the atoms
of the molecule are referred to as the tertiary structure. Quater-nary
structure refers to the arrangement of subunits in proteins consist-ing of more
than one polypeptide chain.
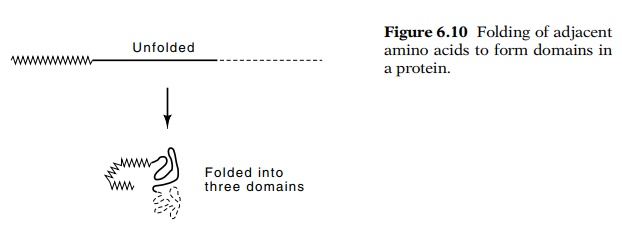
A domain of a protein is a structure unit
intermediate in size between secondary and tertiary structures. It is a local
group of amino acids that have many fewer interactions with other portions of
the protein than they have among themselves (Fig. 6.10). Consequently, domains
are independent folding units. Interestingly, not only are the amino acids of a
domain near one another in the tertiary structure of a protein, but they
usually comprise amino acids that lie near one another in the primary structure
as well. Often, therefore, study of a protein’s structure can be done on a
domain-by-domain basis. The existence of semi-inde-pendent domains should
greatly facilitate the study of the folding of polypeptide chains and the
prediction of folding pathways and struc-tures.
Particularly useful to the ultimate goal of
prediction of protein structure has been the finding that many alterations in
the structure of proteins produced by changing amino acids tend to be local.
This has
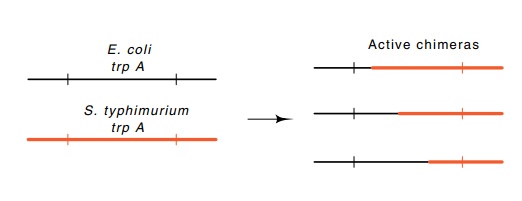
Figure
6.11 Substitution of portions of theE.colitryptophan
synthetaseαsubunit with corresponding
regions from the Salmonella typhimurium
syn-thetase subunit.
been found in exhaustive genetic studies of the lac and lambda phage repressors, in the
thermodynamic properties of mutant proteins, and in the actual X-ray or NMR determined
structures of a number of proteins. In the lac
and lambda repressors, the majority of the amino acid changes that alter the
ability to bind to DNA lie in the portion of the protein that makes contact
with the DNA. Similar results can be inferred from alterations in the amino
acid sequence of the tryptophan synthetase protein generated by fusing two
related but nonidentical genes. Despite appreciable amino acid sequence
differences in the two parental types, the fusions that contain various amounts
of the N-terminal sequence from one of the proteins and the remainder of the
sequence from the other protein retain enzymatic activity (Fig. 6.11). This
means that the amino acid alterations generated by formation of these chimeric
pro-teins do not need to be compensated by special amino acid changes at
distant points in the protein.
The results obtained with repressors and tryptophan
synthetase mean that a change of an amino acid often produces a change in the
tertiary structure that is primarily confined to the immediate vicinity of the
alteration. This, plus the finding that protein structures can be broken down
into domains, means that many of the potential long-range interactions between
amino acids can be neglected and interactions over relatively short distances
of up to 10 Å play the major role in determining protein structure.
The proteins that bind to enhancer sequences in
eukaryotic cells are a particularly dramatic example of domain structures in
proteins. These proteins bind to the enhancer DNA sequence, often bind to small
molecule growth regulators, and activate transcription. In the glucocor-ticoid
receptor protein, any of these three domains may be inde-pendently inactivated
without affecting the other two. Further, domains may be interchanged between
enhancer proteins so that the DNA-bind-ing specificity of one such protein can
be altered by replacement with the DNA-binding domain from another protein.
As we saw earlier in discussing mRNA splicing, DNA regions encod-ing different domains of a protein can be appreciably separated on the chromosome. This permits different domains of proteins to be shuffled so as to accelerate the rate of evolution by building new proteins from new assortments of preexisting protein domains. Domains rather than amino acids then become building blocks in protein evolution.
Related Topics