Chapter: Genetics and Molecular Biology: Transcription,Termination, and RNA Processing
RNA Termination
Termination
Simple experiments modifying the beginning and
middle parts of genes show that most often it is just the sequence at the end
of the transcribed region that specifies transcription termination. One simple
mechanism for transcription termination in bacteria utilizes just the RNA
polym-erase and requires only a special sequence near the 3’ end of the RNA.
The termination signal consists of a region rich in GC bases that can form a
hairpin loop closely followed by a string of U’s. This class of terminators
functions without the need for auxiliary protein factors from the cell.
Frequently termination in eukaryotic systems occurs in a

Most likely as the RNA is elongated past the region
rich in GC, it base pairs with itself to form a hairpin (Fig. 5.5). This
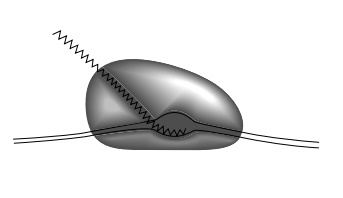
hairpin may fit so poorly in the transcript groove or canyon in the polymerase
(Fig. 5.6), that it weakens the binding of polymerase to the transcription
bubble and also causes the RNA polymerase to pause in this region. Release then
occurs from the run of U’s. Direct physical measurements have shown that oligo
(rU:dA) hybridizes with exceptional weakness com-pared to other
oligonucleotides. The combination of these factors changes the RNA elongation
complex from being extremely stable to being so unstable that transcription
termination usually occurs.
Figure 5.6 The melted DNAand RNA likely fit within can-yons on RNA polymerase. The walls, however, may close in after polymerase binds to DNA to help hold onto the DNA.
A second class of prokaryotic terminators is much
different. This class requires the presence of the rho protein for termination.
The termination activity is further stimulated by a second protein, the nusA gene product. During transcription,
RNA polymerase pauses near a termination sequence, most likely aided in pausing
by the NusA protein, and then rho terminates the transcription process and
releases the RNA and RNA polymerase. Analysis of the 3’ ends of rho-dependent
tran-scripts reveals them to contain no discernible sequence patterns or
significant secondary structures. They have even less secondary struc-ture than
expected for completely random sequences.
Most likely, when the nascent RNA extending from
the RNA polym-erase is free of ribosomes and lacks significant secondary
structure, the
Figure
5.7 RNA hybridized to a circular DNA
molecule is a substrate for rhoprotein plus ATP which separates the two nucleic
acids.
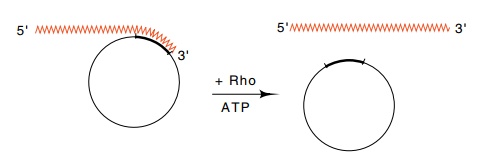
rho protein can bind and move with the consumption
of energy along the RNA up to the polymerase. When it reaches the polymerase,
it separates the growing transcript from the template and terminates
transcription. The separation of the two strands is accomplished by an RNA-DNA
helicase (Fig. 5.7).
The discovery of rho factor was accidental.
Transcription of lambda phage DNA in an in
vitro system produced a large amount of incorrect transcript. This
inaccuracy was revealed by hybridizing the RNA to the two separated strands of
lambda phage. Correct transcripts would have hybridized predominantly to only
one strand. Apparently the conditions being used for transcription did not
faithfully reproduce those existing within the cell and the rho factor somehow
reduced the amount of incorrect transcription. This is a biochemist’s dream for
it means that something must exist and is waiting to be found. Therefore
Roberts looked for and found a protein in cell extracts that would enhance the
fidelity of in vitro transcription.
Upon completing the purification and in studying the properties of his
“fidelity” factor, he discovered that it terminated transcription. Rho factor
shares suggestive properties with a DNA helicase required in DNA replication,
DnaB. Both bind to nucleic acid and move along the nucleic acid with the
consumption of ATP. In the process of this movement, a complementary strand can
be displaced. Further, both helicases are hexameric, and both hydrolyze
significant amounts of ATP when in the presence of a single-stranded
oligonu-cleotide.
Although transcription termination and its
regulation is usually de-termined only by events occurring near the 3’ end of
the transcript, sometimes the 5’ end is also involved. The transcription in E. coli
of ribosomal RNA and of some of the genes of phage lambda depends upon
modifying the transcription complex shortly after the polymerase crosses a
special sequence near the promoter. At this point several proteins bind to the
RNA copy of the sequence and bind to the RNA polymerase as well. After such a
modification, elongation will proceed to the end of the transcription unit and
ignore some opportunities for termination that would be utilized by an unmodified
RNA polymerase. It would be amazing if eukaryotic cells did not also utilize
this as a mechanism for gene regulation.
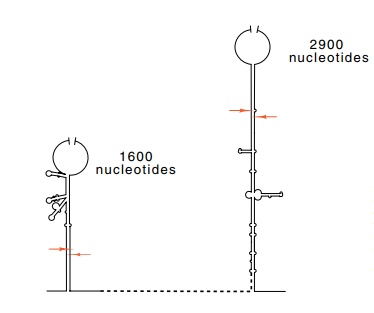
Figure
5.8 Schematic of the basepaired stems
surrounding the 16S and 23S ribosomal RNAs. The ar-rows mark the points of
RNAse III cleavage.
Related Topics