Chapter: Biochemistry: Nucleic Acid Biotechnology
Purification and Detection of Nucleic Acids
Purification and Detection of
Nucleic Acids
In early
1997, headlines around the world reported the successful cloning of a sheep by
Scottish scientists, followed at intervals by reports of cloning of other
animals. Such striking examples of the power of the techniques for manipulating
DNA sparked enormous amounts of discussion.
Experiments
on nucleic acids frequently involve extremely small quantities of materials of
widely varying molecular size. Two of the primary necessities are to separate
the components of a mixture and to detect the presence of nucleic acids.
Fortunately, powerful methods exist for accomplishing both goals.
How are nucleic acids separated?
Separation Techniques
Any separation method depends on the differences between the items to be separated. Charge and size are two properties of molecules that are frequently used for separation. One of the most widely used techniques in molecular biology, gel electrophoresis, uses both these properties. Electrophoresis is based on the motion of charged particles in an electric field. For our purposes, it is enough to know that the motion of a charged molecule in an electric field depends on the ratio of its charge to its mass. A sample is applied to a supporting medium. With the use of electrodes, an electric current is passed through the medium to achieve the desired separation. Polymeric gels, such as agarose and polyacrylamide, are frequently used as supporting media for electrophoresis (Figure 13.1). They are prepared and cast as a continuous cross-linked matrix. The cross-linking gives rise to pores, and the choice of agarose versus polyacrylamide gels depends on the size of the molecules to be separated-agarose for larger fragments (thousands of oligonucleotides) and polyacrylamide for smaller (hundreds of oligonucleotides).
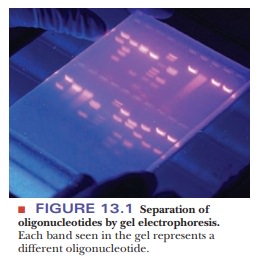
The
charge on the molecules to be separated leads them to move through the gel
toward an electrode of opposite charge. Nucleic acids and oligonucle-otide
fragments are negatively charged at neutral pH because of the presence of the
phosphate groups. When these negatively charged molecules are placed in an
electric field between two electrodes, they all migrate toward the posi-tive
electrode. In nucleic acids, each nucleotide residue contributes a negative
charge from the phosphate to the overall charge of the fragment, but the mass
of the nucleic acid or oligonucleotide increases correspondingly. Thus, the
ratio of charge to mass remains approximately the same regardless of the size
of the molecule in question. As a result, the separation takes place simply on
the basis of size and is due to the sieving action of the gel. In a given
amount of time, with a sample consisting of a mixture of oligonucleotides, a
smaller oligonucleotide moves farther than a larger one in an electrophoretic
separa-tion. The oligonucleotides move in the electric field because of their
charges; the distances they move in a given time depend on their sizes.
Most
separations are done with an agarose gel in a horizontal position, called a submarine gel because it is underneath
the buffer in the chamber. However, when DNA sequencing is done, a
polyacrylamide gel is run in a vertical position. Many different samples can be
separated on a single gel. Each sample is loaded at a given place (a distinct
well) at the negative-electrode end of the gel, and an electric current flows
until the separation is complete (Figure 13.2).

Detection Methods
After
the DNA pieces have been separated, they must be treated in some way that
allows them to be seen. Some of these techniques allow all of the DNA to be
seen, but others are more specific for certain DNA pieces.
How can we visualize DNA?
The
original method for detecting the separated products is based on radioactive
labeling of the sample. A label, or tag, is an atom or molecule that allows
visualization of another molecule. The isotope of phosphorus of mass number 32
(32P, spoken as “P-thirty-two”) was widely used in the past for this
purpose. More recently, 35S, or the isotope of sulfur
of mass number 35 (spoken as “S-thirty-five”), has been used extensively. The
DNA molecules undergo a reaction that incorporates the radioactive isotope into
the DNA. When the labeled oligonucleotides have been separated, the gel is
placed in contact with a piece of X-ray film. The radioactively labeled
oligonucleotides expose the portions of the film with which they are in
contact. When the film is developed, the positions of the labeled substances
show up as dark bands. This technique is called autoradiography, and the resulting film image is an autoradiograph (Figure 13.3).
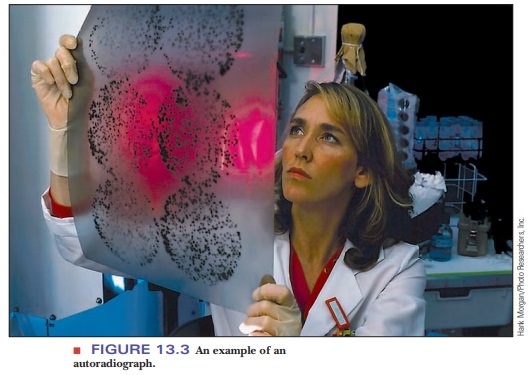
Many examples of autoradiographs can be seen in the scientific literature, but, as time goes on, autoradiography is being replaced by detection methods that do not use radioactive materials and their associated hazards. Many of these methods depend on emission of light (luminescence) by a chemical label attached to the fragments, and they can detect amounts of substances measured in picomoles. The way in which the label emits light depends on the application. When the base sequence of DNA is to be determined, the label is a series of four fluorescent compounds, one for each base. The gel with the separated products is irradiated with a laser; the wavelength of the laser light is absorbed by each of the four labels. Each of the four labeled compounds re-emits light at a different, characteristic, longer wavelength. This is called fluorescence. Another detection method that uses fluorescence involves the compound ethidium bromide.
Its molecular structure includes a planar portion
that can slip between the bases of DNA, giving ethidium bromide fluores-cent
properties when it binds to DNA that differ from those observed when it is free
in solution. An ethidium bromide solution is used as a stain for DNA in a gel.
The solution soaks into the gel, and the DNA fragments in the gel can be seen
as orange bands by shining ultraviolet light on the gel.
Summary
In order to study nucleic acids, we must have a way to separate and
iden-tify them.
The most common separation technique is gel electrophoresis. DNA
pieces separate on a gel based on size.
DNA can be visualized on a gel via autoradiography, a procedure in
which the DNA has incorporated radioactive nucleotides.
DNA can
also be seen using fluorescence, often with a compound called ethidium bromide,
which glows orange under UV light when bound to DNA.
Related Topics