Chapter: Plant Biochemistry: Protein biosynthesis occurs in three different locations of a cell
Proteins attain their three-dimensional structure by controlled folding
Proteins attain their three-dimensional structure by controlled folding
Protein biosynthesis by ribosomes first yields an unfinished (immature) polypeptide. This is converted (if necessary after amino acid sequences, e.g., transit or signal sequences are cleaved off) by specific folding to establish the biological active form, the native protein. The three-dimensional structure of the native protein normally represents the lowest energy state of the molecule and is determined to a large extent by the amino acid sequence of the molecule.
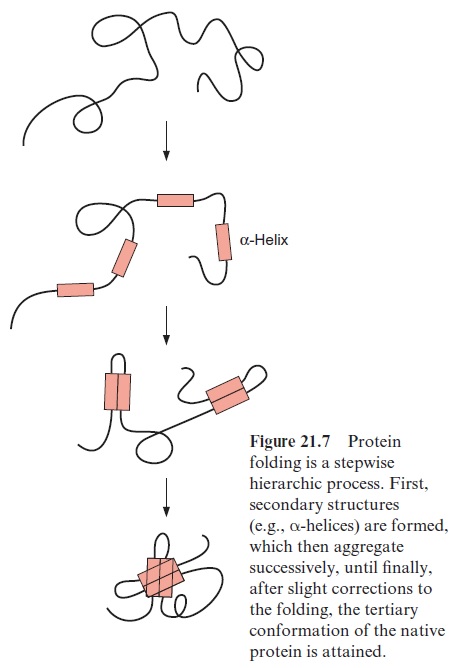
The folding of a protein is a multistep process
Theoretically there are about 10100 possible conformations for a peptide with 100 amino acids, which is a rather small protein of about 11 kDa. Since the reorientation of a single bond requires about 10–13 s, it would take the incred-ibly long time of 1087 s to try all possible folding states one after the other. By comparison, the age of the earth is about 1.6 1017 s. In reality, a protein attains its native form within seconds or minutes. Apparently the folding of the molecule proceeds in a multistep process. It begins by forming second-ary structures such as α-helices or β-sheets. They consist of 8 to 15 amino acid residues, which are generated or disintegrated within milliseconds (Fig. 21.7). The secondary structures then associate stepwise with increasingly larger domains and in this way also stabilize the regions of the molecule that do not form secondary structures. The driving force in these folding processes is the hydrogen bond interactions between the secondary struc-tures. After further conformational changes, the correct three-dimensional structure of the molecule is attained rapidly by this cooperative folding pro-cedure. In proteins with several subunits, the subunits associate to form a quaternary structure.
Proteins are protected during the folding process
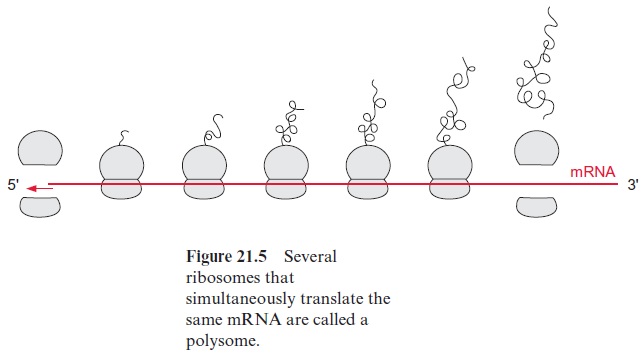
The folding process can be severely disturbed when the secondary struc-tures in the molecule associate incorrectly, or particularly when second-ary structures of different molecules associate, resulting in an undesirable aggregation of proteins (hydrophobic collapse). This danger is especially high during protein synthesis, when the incomplete protein is still attached to the ribosome (Fig. 21.5), or during the transport of an unfolded pro-tein through a membrane, when only part of the peptide chain has reached the other side. Moreover, incorrect intermolecular associations are likely to occur when the concentration of a newly synthesized protein is very high, as can be the case in the lumen of the rough endoplasmatic reticu-lum . To prevent such incorrect folding, a family of proteins present in the various cell compartments helps newly formed protein molecules to attain their correct conformation by avoiding incorrect asso-ciations. These proteins have been named chaperones.
Heat shock proteins protect against heat damage
Chaperones not only have a function during correct folding of proteins but also protect proteins against aggregation when they have been dena-tured by exposure to high temperatures, thus assisting their reconversion to the native conformation. Bacteria, animal, and plant cells react to a temperature increase of about 10% above the temperature optimum with a very rapid synthesis of so-called heat shock proteins, most of which are chaperones. Many plants can survive otherwise lethal high temperatures if they have been pre-exposed to a smaller temperature increase, which had induced the synthesis of heat shock proteins. This phenomenon is called acquired thermal tolerance. Investigations with soybean seedlings showed that such tolerance coincides with an increase in the content of heat shock proteins. However, most of these heat shock proteins are constitutively present in the cells, which indicates that heat shock proteins have impor-tant functions in the folding of proteins even under normal conditions.
Chaperones bind to unfolded proteins
Since chaperones were initially characterized as heat shock proteins, they are commonly designated by the abbreviation Hsp followed by the molecu-lar mass in kDa.
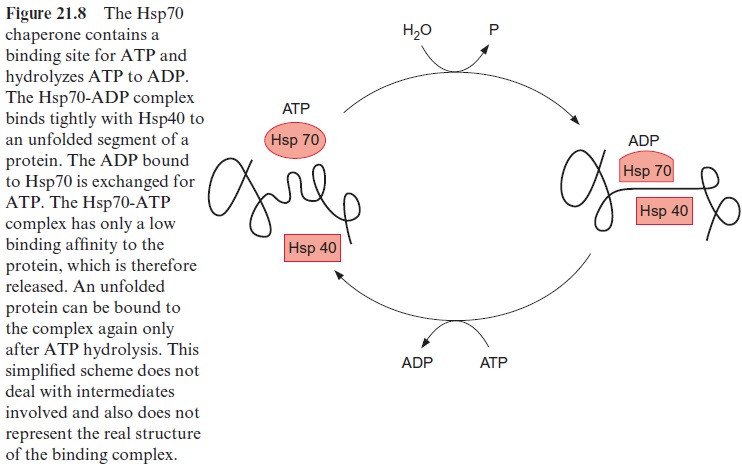
Chaperones of the Hsp70 family have been found in bacteria, mitochon-dria, chloroplasts, and the cytosol of eukaryotes, as well as in the endoplas-matic reticulum. These are highly conserved proteins. Hsp70 has a binding site for adenine nucleotides, which can be occupied either by ATP or ADP. When occupied by ADP, Hsp70 forms with the chaperone Hsp40 (also required for the binding of unfolded proteins) a tight complex with unfolded segments of a protein, but not with native proteins (Fig. 21.8). The ADP bound to Hsp70 is subsequently replaced by ATP. The resultant ATP-Hsp70 complex has only a low binding affinity and therefore dissociates from the protein segment. Due to the subsequent hydrolysis of the bound ATP to ADP, Hsp70 is ready to bind once more to an unfolded peptide segment. In this way Hsp70 binds to a protein only for a short time, dissociates from it, and, if necessary, binds to the protein again. This stabilizes an unfolded protein without restricting its folding capacity. The mechanism of the ADP-dependent binding of Hsp70 to unfolded peptides as mediated by Hsp40 has been conserved during evolution. Fifty percent of the amino acids in the sequences of the Hsp70 protein in E. coli and in humans are identical.
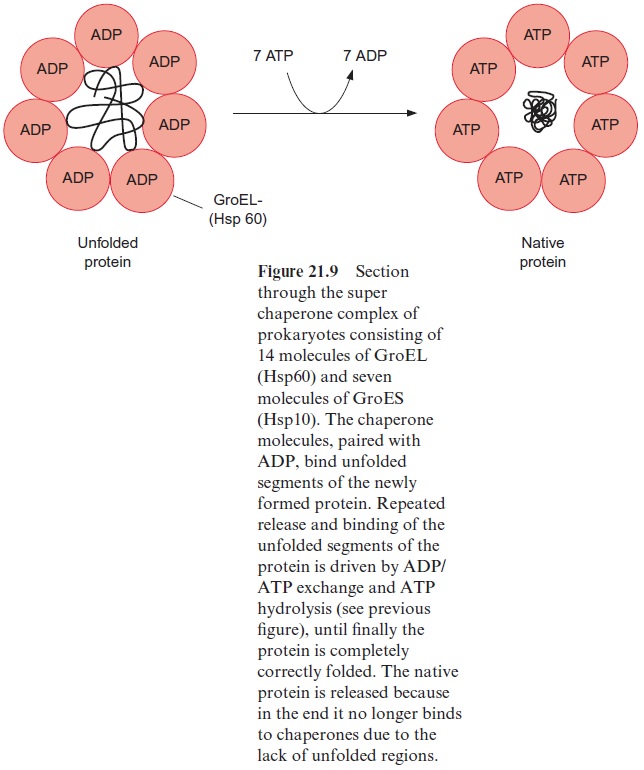
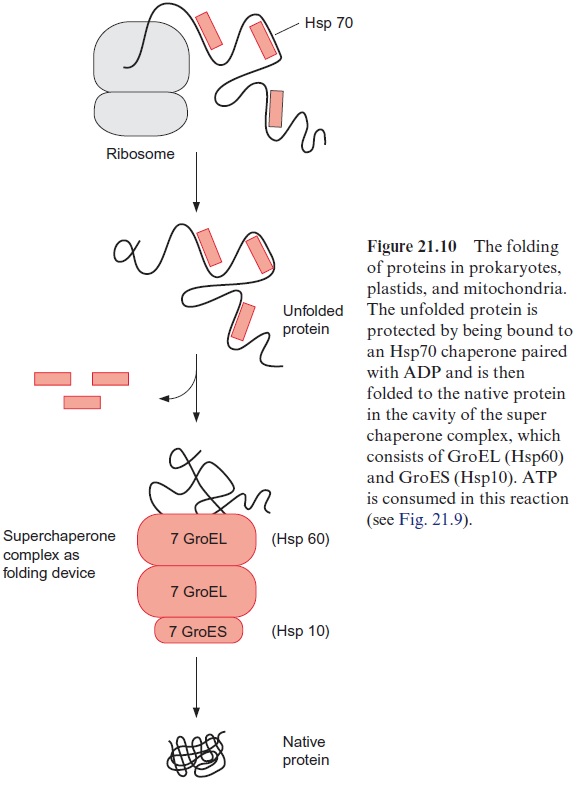
The proteins of the Hsp60 family, present in bacteria, plastids, and mito-chondria, also bind to unfolded proteins. They were first identified as the GroEL factor in E. coli and as RubisCO binding protein in chloroplasts, until it was realized that both proteins are homologous and act as chaperones. A GroES factor called Hsp10 in mitochondria and in chloroplasts is involved in binding the Hsp60 chaperones. In bacteria, 14 GroEL and 7 GroES mol-ecules are assembled to a superchaperone complex, forming a large cavity into which an unfolded protein fits (Figs. 21.9, 21.10). The unfolded protein is temporarily bound to Hsp60 molecules of the cavity analogously to the bind-ing to Hsp70 in Figure 21.8. Correct folding to the native protein is aided by several ATP hydrolysis cycles, involving dissociation and rebinding of the protein segments. In this way the unfolded protein can reach the native con-formation by avoiding association with other proteins.
Hsp70 as well as Hsp60 and Hsp10 participate in the protein folding in plastids and mitochondria (Fig. 21.10). Hsp70 protects single segments of the growing peptide chain during protein synthesis, and the super chaperone complex from Hsp60 and Hsp10 finally enables the undisturbed folding of the total protein. The chaperone Hsp90 is found in very high concentrations in the cytosol of eukaryotes. Hsp90 with co-chaperone (HOP) is regarded as playing a central role in the folding and assembling of cytosolic pro-teins. Moreover, chaperones named CCT (cytosolic complex T), somewhat resembling the prokaryotic Hsp60, have been identified in the cytosol of eukaryotes. They act as a folding device by forming oligomeric chaperone complexes, which are probably similar to the Hsp60-Hsp10 super chaper-one complex (Fig. 21.10).
Furthermore, there are proteins facilitating other processes, which limit protein folding, such as the formation of disulfide bridges and the cis-trans isomerization of the normally non-rotatable prolyl peptide bonds. Since thorough investigation of chaperones began only a few years ago, many questions about their structure and function are still unanswered.
Related Topics