Chapter: Biochemistry: Proteins
Protein structure
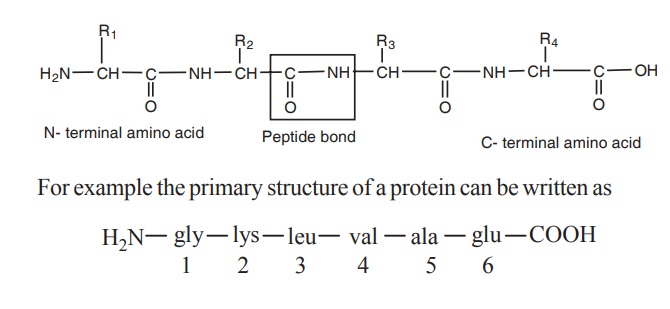
Protein structure
The architecture of protein molecule is complex
but well organised. To understand this, a clear idea of certain basic details
regarding the mode of arrangement of the structural units inside the molecule
is necessary. Linder strom - Lang suggest four types of structural organisation
for proteins. They are
1. Primary structure
2. Secondary structure
3. Tertiary Structure and
4. Quarternary strucutre
1. Primary structure
The primary structure of protein is defined as
the sequence of amino acid residues making up its polypeptide chain. The
protein may be formed of one or more polypeptide chains. The amino acids are
arranged in specific sequence in these polypeptide chain. The amino acid
residues are linked by peptide bonds. The peptide bond is formed between the
carboxyl group of one amino acid and the amino group of adjacent amino acid.
Some times the adjacent polypeptide chains are linked by disulphide bonds.
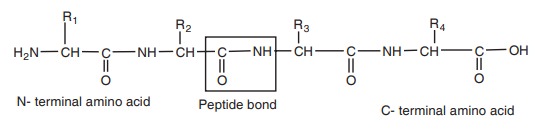
For example the primary structure of a protein can be written as

Each polypeptide chain of any length has at one
end a N-terminal amino acid containing free amino group and at the other end a
C-terminal amino acid containing a free carboxyl group. The amino acids in a
polypeptide chain are numbered from the N-terminal end
The primary structure has the following salient
features
·
Primary
structure refers to the linear sequence of amino acid residues.
·
The
proteins are linear and unfolded
·
The
protein is formed of one or more polypeptide chains.
·
The
amino acid residues are linked by repeating polypeptide bonds.
·
The
adjacent polypeptide chains are linked by disulphide bonds.
·
Most of
the structural proteins which are in the form of fibres exhibit primary
structure
·
The
primary structure provides information on the number and proportion of
different amino acids in a protein. Primary structures of a large number of
proteins have been determined.
eg.
i. human insulin has 51 amino acids distributed
in two poly peptide chains. A chain-31 amino acids, B chain-20 amino acids and
the polypeptides are linked by disulphide bridges.
ii. cytochrome C contains 104 amino acids.
iii. human serum albumin contains 584 amino
acids.
Primary structure is ultimately responsible for
the native structure of the protein.
2. Secondary structure
The peptide chain thus formed assumes a
two-dimentional secondary structure by way of folding or coiling consisting of
a helically coiled, zig-zag linear or mixed form. It results from the steric
relationship between amino acids located relatively near to each other in the
peptide chain. The linkages or bonds involved in the secondary structure
formation are hydrogen bonds and disulphide bonds.
i. Hydrogen bond
These are weak, low energy non-covalent bonds sharing a single hydrogen by two electronegative atoms such as O and N. Hydrogen bonds are formed in secondary structure by sharing H-atoms between oxygen of
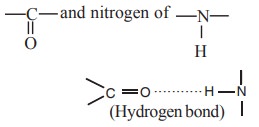
of different peptide bonds.
The hydrogen bonds in secondary structure may
form either an α- helix or β-pleated sheet structure.
ii. Disulphide bond
These are formed between two cysteine residues.
They are strong, high energy covalent bonds.
Proteins exist in the two forms of secondary
structure, a helix and b pleated sheet
α- helix
A polypeptide chain forms regular helical coils
called α- helix. These coils are stabilized by hydrogen bonds between carbonyl
oxygen of first amino and amide N of fourth amino acid residues. Thus in α- helix intra chain hydrogen bonding is present. The a-helices can be
either right handed or left handed. Left handed α-
helix is less stable because of the steric
interference between the carbonyl group and the side chains. Only the right
handed α- helix has been found in protein structure (Fig.5.5).
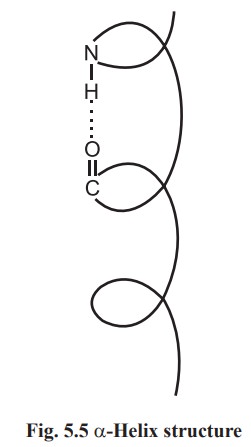
Each amino acid residue advances by 0.15 nm
along the helix and 3.6 amino acid residues are present in one complete turn.
The distance between two equivalent points on turn is 0.54 nm and is called a
pitch.
Small or uncharged amino acid residues such as
alanine, leucine and phenyl alanine are often found in α- helix. More polar residues such as arginine, glutamate and serine may
repel and destabilize α- helix. Proline is never found in α- helix.
Hair, nail, skin contain a group of proteins
called keratins rich in α - helical structure.
beta-pleated sheet structure
A conformation called b pleated sheet structure is thus formed when hydrogen bonds are
formed between the carbonyl oxygens and amide hydrogens of two or more adjacent
extended polypeptide chains. Thus the hydrogen bonding in b pleated sheet structure is interchain. The structure is not
absolutely planar but is slightly pleated due to the bond angles. The adjacent
chains in β-pleated sheet structure are either parallel or antiparallel, (Fig.5.6)
depending on whether the amino to carbonyl peptide linkage of the chains runs
in the same or opposite direction.
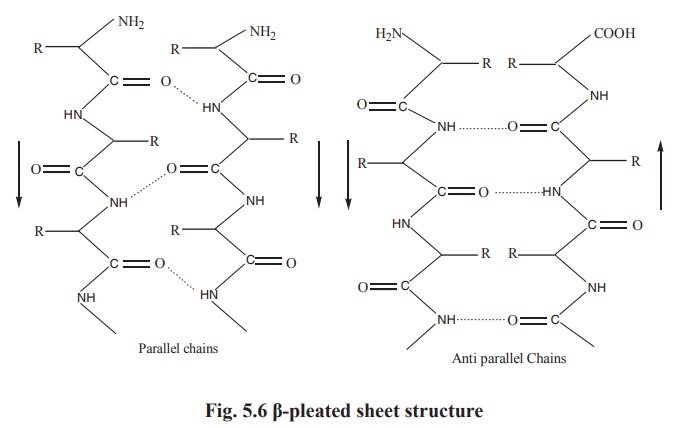
In both parallel and antiparallel β-pleated sheet structures, the side chains are on opposite sides of the
sheet. Generally glycine, serine and alanine are more common to form β-pleated sheet. Proline occurs in β-pleated sheet although it tends to distrupt the sheets
by producing links. Silk fibroin, a protein of silk worm is rich is β-pleated sheet.
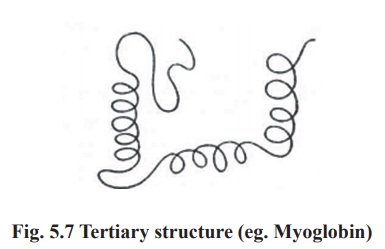
3. Tertiary structure
The polypeptide chain with secondary structure
may be further folded, super-folded, twisted about itself forming many sizes.
Such a structural confirmation is called tertiary structure. It is only one
such confirmation which is biologically active and protein in this conformation
is called as native protein. Thus the tertiary is constitued by steric
relationship between the amino acids located far apart but brought closer by
folding (Fig. 5.7). The bonds responsible for interaction between groups of
amino acids are as follows.
i. Hydrophobic interactions
Normally occur between nonpolar side chains of
amino acids such as alanine, leucine, methionine, isoleucine and phenyl
alanine. They constitute the major stabilzing forces for tertiary structure
forming a compact three-dimentional structure.
ii. Hydrogen bonds
Normally formed by the polar side chains of the
amino acids.
iii. Ionic or electrostatic interactions
The interaction occurs between oppostively
charged polar side chains of amino acids, such as basic and acidic amino acids.
iv. Vander -wall forces
Occurs between non polar side chains.
v. Disulphide bonds
These are S-S bonds formed between - SH groups
of distant cysteine residues.
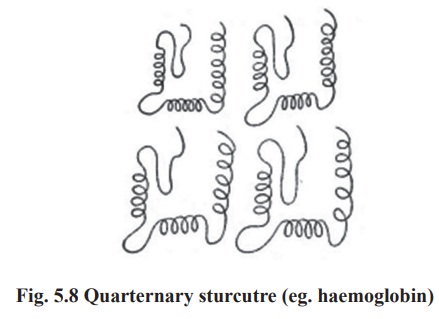
4. Quarternary structure
Some proteins are made up of more than one
polypeptide chain These peptide chains held together by non-covalent
interactions or by covalent cross - links it is referred to as the quarternary
structure. The assembly is often called as an oligomer and each constituent
peptide chain is called as a monomer or sub unit. The monomers of oligomeric
protein can be identical or quite different in primary, secondary or tertiary
structure (Fig. 5.8).eg:
Proteins with 2 monomers (dimer) eg. Creatine
phosphokinase
Proteins with 4 monomers (tetramer) eg.
Haemoglobin
Related Topics