Chapter: Biotechnology Applying the Genetic Revolution: Proteomics
Protein Interactions: The Yeast Two Hybrid System
PROTEIN
INTERACTIONS: THE YEAST TWO-HYBRID SYSTEM
In addition to protein
function and expression, proteomics attempts to find relevant protein
interactions. For those who like “-omics” terminology, the total of all
protein-protein interactions is called the protein
interactome. For example, hormones usually bind to receptors that pass the
signal on. Often this involves a protein relay where one protein activates
another, which in turn activates yet another. To understand hormone function,
researchers must identify all the proteins in the signal cascade. Phage display
is one way to identify interactions, but the displayed proteins may not fold
correctly or specific cofactors may be missing when mammalian proteins are
expressed in bacteria.
An approach to overcoming
these difficulties is the yeast two-hybrid
system, where the binding of two proteins activates a reporter gene. The
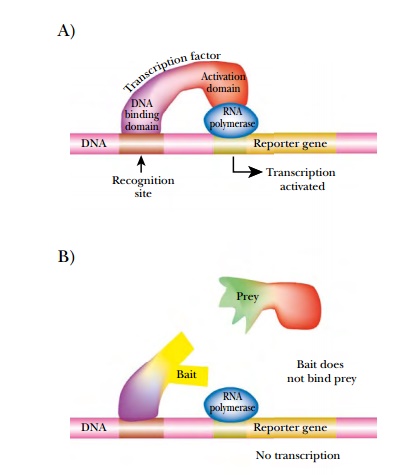
binding of a transcriptional activator protein, GAL4, to the promoter region of
the reporter gene activates transcription and translation. GAL4 contains two
domains needed to turn on the reporter gene. The DNA binding domain (DBD)
recognizes the promoter element and positions the second domain, the activation
domain (AD) next to RNA polymerase, where it activates transcription. These two
domains can be expressed as separate proteins, but cannot activate the reporter
gene unless they are brought together (Fig. 9.21).
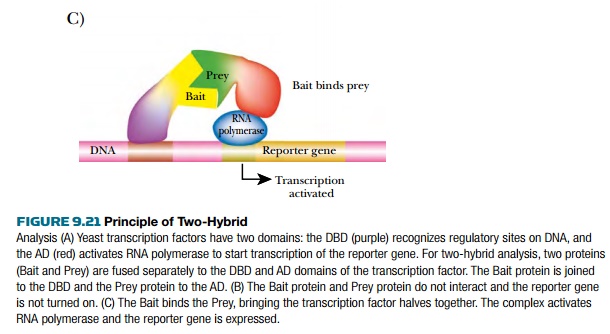
In the two-hybrid system, the
two domains are each fused to different proteins by creating hybrid genes. The bait is the DBD genetically fused to
the protein of interest, and the prey
is the AD fused to proteins that are being screened for interaction with the
bait. When the bait and prey bind, the DBD and AD activate transcription of the
reporter gene.
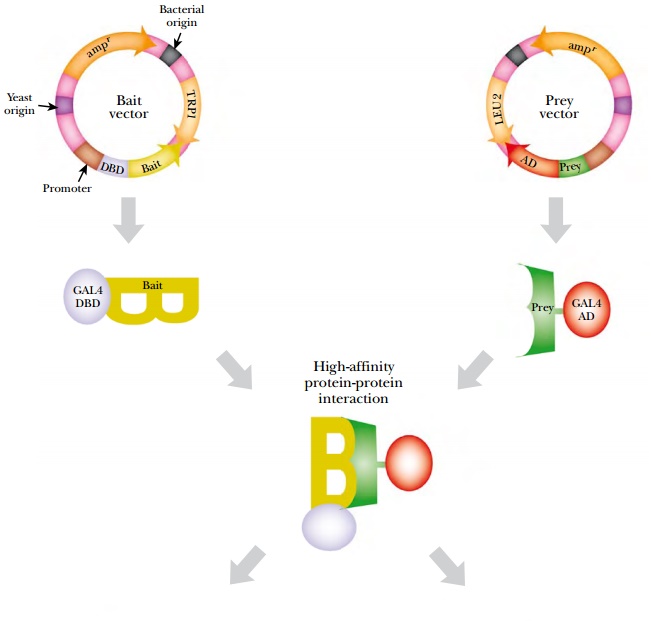
Two vectors are needed to perform two-hybrid analysis (Fig. 9.22). The first vector has a multiple cloning site for the bait protein 3′ of the GAL4-DBD; therefore, the fusion protein has the Bait protein as its C-terminal domain. The second vector has a multiple cloning site for the Prey protein 5′ of GAL4-AD and the fusion protein has the Prey as its N-terminal domain. Both plasmids must be expressed in the same yeast cell. If the bait and prey proteins interact, the reporter gene is turned on.
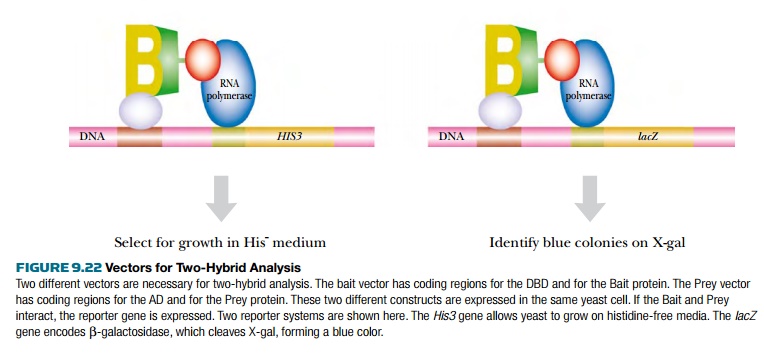
The reporter genes must be
engineered to be under control of the GAL4 recognition sequence. Common
reporter genes include HIS3, which
encodes an enzyme in the histidine pathway and whose expression allows yeast
cells to grow on media lacking histidine, or URA3, which allows growth without uracil. These reporter systems
require yeast host cells that are defective in the corresponding genes.
However, they do allow direct selection of positive isolates.
Another reporter used is lacZ from E. coli, which encodes β-galactosidase. Both bacteria and yeast that
express lacZ turn blue when grown
with X-Gal. β-galactosidase cleaves X-Gal,
releasing a blue product. The reporter genes are usually integrated into the
yeast genome, rather than being carried on a separate vector.
The yeast two-hybrid system
has been used to identify all the protein interactions in the yeast proteome by
mass screening with mating (Fig. 9.23). Yeast has about 6000 different
proteins, and each of these has been cloned into both vectors via PCR. This
way, each protein can be used as both bait and prey. All the bait vectors were
transformed into haploid yeast of one
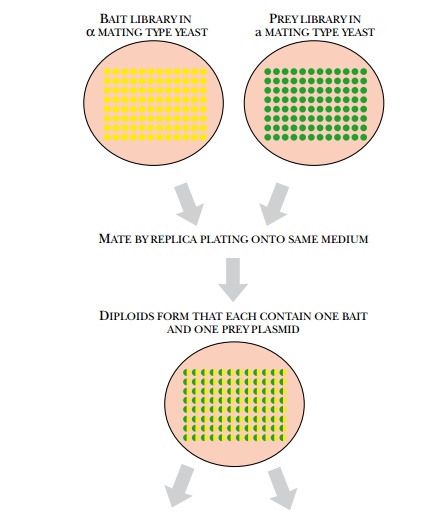
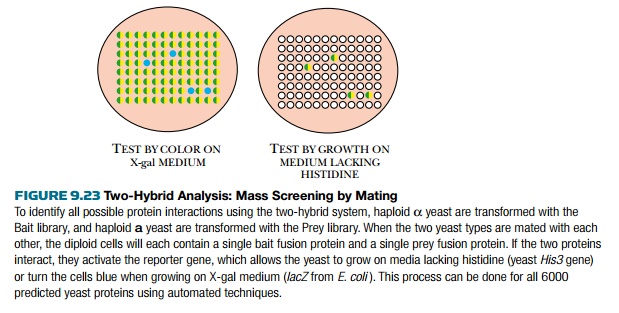
mating type, and the prey
vectors into the other mating type. Haploid cells carrying bait are fused to
haploid cells with prey and the resulting diploid cells are screened for
reporter gene activity. This analysis thus examined 6000 × 6000 combinations for protein interaction.
The yeast two-hybrid system
has some limitations. Because transcription factors must be in the nucleus to
work, the target proteins must also function in the nucleus. For some proteins,
entering the nucleus may cause the protein to misfold. For other proteins, the
nucleus does not have the proper cofactors and the protein may be unstable.
Large proteins may not be expressed well, or may be toxic to the yeast, leading
to false negative results.
Related Topics