Chapter: Biochemistry: Protein Synthesis: Translation of the Genetic Message
Protein Degradation
Protein Degradation
One of
the most often overlooked controls of gene expression occurs at the level of
the degradation of proteins. Proteins are in a dynamic state in which they are
turned over often. Athletes are painfully aware of this because it means that
they must work very hard to get in shape, but then they get out of shape very
quickly. Some classes of proteins experience a 50% turnover every three days.
In addition, abnormal proteins that were formed from errors in either
transcription or translation are degraded quickly. It is believed that a single
break in the peptide backbone of a protein is enough to trigger the rapid
degradation of the pieces, because breakdown products from natural proteins are
rarely seen in vivo.
How does the cell know which proteins to degrade?
If
protein degradation is so quick, clearly it is a process that must be heavily
controlled to avoid destruction of the wrong polypeptides. The degradation
pathways are restricted to degradative subcellular organelles, such as
lysosomes, or to macromolecular structures called proteasomes. Proteins are directed to lysosomes by specific signal
sequences, often added in a posttranslational modification step. Once in the
lysosome, the destruction is nonspecific.
Proteasomes are found in both prokaryotes and eukaryotes, and specific pathways exist to target a protein so that it complexes with a proteasome and is degraded.
In eukaryotes, the most common mechanism for targeting protein for destruction in a proteasome is by ubiquitinylation. Ubiquitin is a small polypeptide (76 amino acids) that is highly conserved in eukaryotes. There is a high degree of homology between the sequences in species as widespread as yeast and humans. When ubiquitin is linked to a protein, it condemns that protein to destruction in a proteasome. Figure 12.23 shows the mechanism of ubiquitinylation.
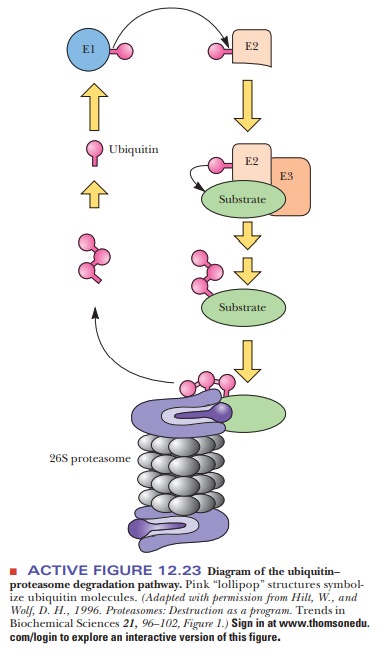
Three enzymes are involved—ubiquitin-activating enzyme (E1), ubiquitin-carrier protein (E2), and ubiquitin-protein ligase (E3). The ligase transfers the ubiquitin to free amino groups on the targeted protein, either the N-terminus or lysine side chains. Proteins must have a free α-amino group to be susceptible, so proteins that are modified at the N-terminus—with an acetyl group, for instance—are protected
from ubiquitin-mediated degradation. The nature of the N-terminal amino acid
also influences its susceptibility to ubiquitinylation. Proteins with Met, Ser,
Ala, Thr, Val, Gly, or Cys at the N-terminus are resistant. Those with Arg,
Lys, His, Phe, Tyr, Trp, Leu, Asn, Gln, Asp, or Glu at the N-terminus have very
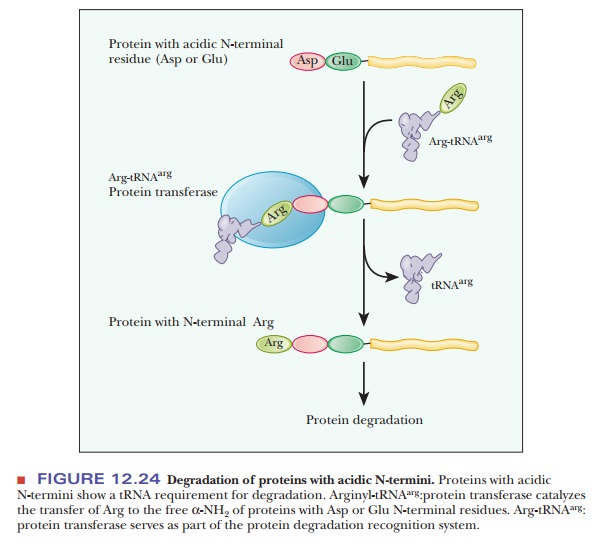
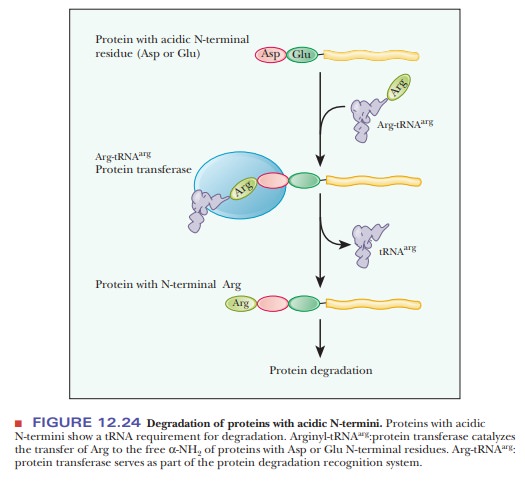
short half-lives, between 2 and 30 minutes. Proteins with an acidic residue at
the N-terminus have a requirement for tRNA as part of their destruction
pathway. The tRNA for arginine, Arg-tRNAarg, is
used to transfer arginine to the N-terminus, making the protein much more
susceptible to the ubiquitin ligase (Figure 12.24). The following Biochemical
Connections box gives an interest-ing example of how transcription regulation
and protein degradation work together to control the process of acclimation to
high altitude.
Summary
Proteins are degraded in subcellular
organelles, such as lysosomes, or in macromolecular structures called
proteasomes.
Many proteins are targeted for destruction by
being bound to a protein called ubiquitin.
The
nature of the amino acid sequence at the N-terminus is often very important to
control of the timing of destruction of a protein.
Related Topics