Chapter: Genetics and Molecular Biology: DNA Synthesis
Proofreading, Okazaki Fragments, and DNA Ligase - Enzymology
Enzymology
Proofreading, Okazaki Fragments, and DNA Ligase
As already discussed, a single strand of DNA
possesses a polarity resulting from the asymmetric
deoxyribose-3’-phosphate-5’-de-oxyribose bonds along the backbone. Most DNA
found in cells is double-stranded; a second strand is aligned antiparallel to
the first strand and possesses a sequence complementary to the first. This
self-complementary structure solves problems in replication because
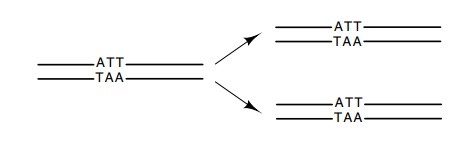
the product of replication is two daughter
molecules, each identical to one of its parents. As a result of the structural
similarity between the parent and daughter duplexes, the mechanisms necessary
for readout of the genetic information or replication of the DNA need not
accom-modate multiple structures. Further, the redundancy of the stored
information permits DNA with damage in one strand to be repaired by reference
to the sequence preserved in the undamaged, complementary strand.
As is often the case in biology, numerous
illustrative exceptions to the generalizations exist. Single-stranded DNA phage
exist. These use a double-stranded form for intracellular replication but
encapsidate only one of the strands. Apparently, what they lose in repair
abilities they gain in nucleotides saved.
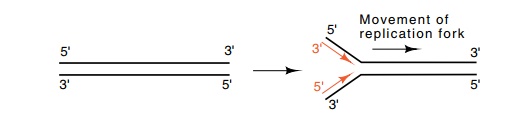
Generally, DNA is double-stranded, and both strands are replicated almost simultaneously by movement of a replication fork down the DNA. In this case, the opposite polarities of the
strands require that as a replication fork moves down the DNA, overall, one
daughter strand grows in a 3’-to-5’ direction and the other daughter strand
grows in a
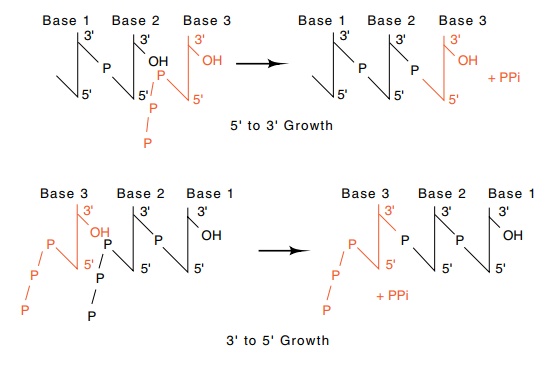
Figure
3.1 DNA could theoretically be
elongated in both the 5’-to-3’and the3’-to-5’ directions with the use of only
nucleoside 5’ triphosphates.
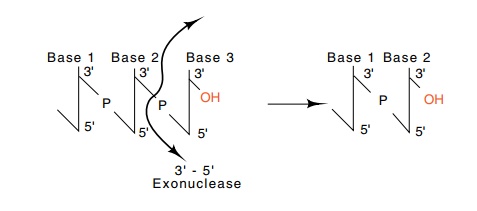
Figure
3.2 Exonucleolytic removal of the
final nucleotide from a chain elon-gating in the 5’-to-3’ direction, a 3’-5’
exonucleolytic activity, regenerates the 3’-OH.
Although cells have been found to possess
5’-nucleoside triphos-phates and not 3’-nucleoside triphosphates, the two
daughter DNA strands could be elongated nucleotide by nucleotide in both the
5’-to-3’, and the 3’-to-5’ directions using just 5’-nucleoside molecules (Fig.
3.1). This latter possibility is largely excluded, however, by the need for
proofreading to check the accuracy of the latest incorporated nucleo-tide.
If the nucleotide most recently incorporated into
the elongating strand does not correctly pair with the base on the
complementary strand, the misincorporated nucleotide ought to be excised.
Ultimately such editing to remove a misincorporated base must generate a DNA
end precisely like the end that existed before addition of the incorrectly
paired nucleotide. Removal of the final nucleotide from the strand growing in
the 5’-to-3’ direction immediately regenerates the 3’-OH that is normally found
at the end (Fig. 3.2). A strand growing in the 3’-to-5’ direction utilizing 5’
triphosphates will possess a triphosphate on its 5’ end. Simple excision of the
final nucleotide from such a strand does not regenerate the 5’ triphosphate
end. Creation of the end normally seen by the polymerase elongating such a
strand would then require another enzymatic activity. This, in turn, would
require dissociation of the polymerase from the DNA and the entry of the other
enzyme, a process that would drastically slow the process of DNA elongation.
It is worthwhile examining why DNA polymerase must
remain bound to the complex of template strand and elongating strand through
thou-sands of elongation cycles. Such a processive behavior is essential since
elongation rates per growing chain must be hundreds of nucleotides per second.
If the polymerase dissociated with the addition of each nucleo-tide, it would
have to bind again for the next nucleotide, but even with moderately high concentrations
of polymerase, the binding rate of a protein to a site on DNA in cells is about
one per second to one per 0.1 second, which is far below the necessary
elongation rate. Consequently, accurate DNA synthesis prohibits elongation in
the 3’-to-5’ direction.
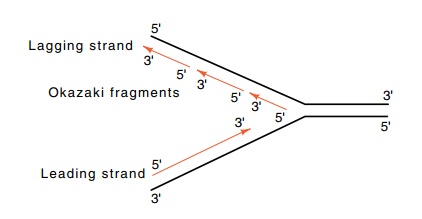
Figure
3.3 Nomenclature of the leading and
lagging strands.
A variety of experiments show that both strands are
synthesized at replication forks. This means that overall, one strand grows in
the 5’-to-3’ direction and the other in the 3’-to-5’ direction. One strand can
be synthesized continuously in the 5’-to-3’ direction. The other strand cannot.
It must grow in the 3’-to-5’ direction by the synthesis of short segments which
themselves are elongated in the 5’-to-3’ direction. These are ligated together
so that the net growth of this strand is 3’-to-5’ (Fig. 3.3). These fragments
are called Okazaki fragments after their discov-erer. The strand synthesized
continuously is called the leading strand, and the discontinuously synthesized
strand is the lagging strand.
The above-mentioned considerations suggest that DNA
polymerases should have the following properties. They should use 5’-nucleoside
triphosphates to elongate DNA strands in a 5’-to-3’ direction, and they should
possess a 3’-to-5’ exonuclease activity to permit proofreading. Additionally,
cells should possess an enzyme to join the fragments of DNA that are
synthesized on the lagging strand. This enzyme is called DNA ligase.
Related Topics