Chapter: Plant Biology : Reproductive ecology
Polymorphisms and population genetics
POLYMORPHISMS AND POPULATION GENETICS
Key Notes
Visible variation
Species may vary geographically or in response to ecological conditions. Some variation is continuous, some discontinuous. When there is discontinuous variation within a population it is known as a polymorphism.
Biochemical variation
Polymorphisms in enzymes can be detected by electrophoresis and many hundreds of plants have been studied. A further range of variation is known through analysis of DNA using restriction fragment length polymorphism (RFLP) and random amplified polymorphic DNA (RAPD) techniques. Some enzymes and DNA fragments do not vary across a wide taxonomic range, but some are highly variable between individuals.
Gene flow
There are two components: pollen flow and seed dispersal, both of which are restricted in all plants. Self-incompatible wind-pollinated plants with good seed dispersal have potentially long-distance gene flow compared with self-fertile plants with poor seed dispersal.
Natural selection
The importance of natural selection in the maintenance of genetic variation is much disputed. Very little of the biochemical variation detected has a known selective basis but it may be hard to detect. Heterozygous plants appear to be at an advantage in many species, indicating that cross-fertilization will be selected for.
Population genetic Structure
The total amount of genetic variation differs greatly between plants and how it is distributed between individuals within each population or between populations. This depends on the breeding system and seed dispersal and if these are restricted then restricted gene flow is the dominating influence. If the plant is self-incompatible with good seed dispersal, frequently the dominant plants, natural selection appears to be more important. Isolated small populations are frequently limited by a small number of founders or random extinction of some morphs.
Visible variation
Individual plants in any one species vary. Marked differences between individuals are most commonly seen as geographical variations or variations in response to ecological conditions. These are known as subspecies if the variation appears to have no obvious ecological basis, or ecotypes if it is aresponse to ecological conditions. Plants are only considered to be ecotypically distinct if the differences have a genetic basis. Plants can respond directly to environmental conditions by longer stems and smaller leaves in dark places for example, but if any differences remain when the plants are grown in uniform conditions it indicates a genetic basis. Many ecotypes are morphological, such as short-growing ecotypes from exposed or heavily grazed places, and some may be physiological, such as salt-tolerant ecotypes near coasts.
Variation may occur within populations too, and some plants show a range of morphological variation. This may be continuous, e.g. in leaf shape, or discontinuous such as variation in flower color or the presence of hairs on the seeds. Some of the variants, or morphs, occur at a low frequency of less than 1 in 20 plants, and are maintained only by the occasional recurrent appearance from mutation or rare gene combination. Many blue or purple-colored flowers have occasional white morphs at low frequency. When morphs occur at greater frequency than 1 in 20, the feature is polymorphic and the plant shows a polymorphism.
Biochemical variation
A great range of polymorphic variation occurs in proteins, most easily detected in enzymes using a technique known as electrophoresis developed in the 1970s (Fig. 1).
Isozymes
Isozymes are genetic variants of enzymes that have differing electrophoretic mobilities, due to changes in overall charge following amino acid substitution or changes in size of the molecule. Measurement is usually made using enzymespecific colorimetric stains. Reliance on measurable changes in mobility means that much variation will go undetected, but the method is robust and fairly cheap. Many hundreds of plant species have been screened in this way and at least a little variation has been detected in almost all sexually reproducing plants. In a variation of this technique, iso-electric focussing, isozymes are focussed to a point of equal charge and migrate according to size.
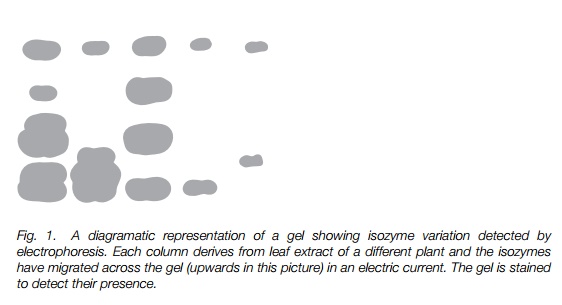
Restriction fragment length polymorphism (RFLP)
Since the 1990s, DNA content has been analyzed directly. The RFLP method relies on the detection of sequence changes in DNA through the use of sequence-specific restriction enzymes isolated from certain bacteria. These cleave DNA at a target site; mutations at this site will prevent cleavage leading to changes in the length of the target DNA. The sites are examined by a DNA hybridization technique known as Southern blotting or by using a polymerasechain reaction (PCR) to amplify the DNA fragments which are then separated by electrophoresis and detected using probes of luminescent or radioactively marked DNA or RNA sequences that bind to the DNA.
Random amplified polymorphic DNA (RAPD)
In techniques involving RAPD, copies of DNA are generated by synthesizing fragments of DNA between two identical short sequences known asprimers (usually 10 bp) using PCR. It is possible to sample many sites in the genome at
once using this technique. Two sites for a single primer may exist in close proximity on a genome and the intervening fragment will be amplified, but for each primer a variable number of such sites will exist so a variable number of different-sized fragments will be produced. These can be analyzed using electrophoresis and the multiplicity of the sites means that different individuals may differ in the fragments amplified. The problems with this technique are that small changes in primer concentration or precise conditions in the experimental set-up can lead to different results and that certain DNA fragments may migrate at the same speed as other unrelated fragments so analysis of the bands can be problematic.
A combination of RFLP and RAPD techniques may be used to give a DNA ‘fingerprint’, used in forensic testing and taxonomic studies as well as in thestudy of variation. Certain stretches of DNA do not vary at all within species orbetween species and sometimes across orders or even kingdoms, whereas othersshow so much variation that every individual is different. The most usefuldifferences for studying variation in plants and their populations are thosebetween these two extremes.
Gene flow
Study of plant polymorphisms has given much insight into gene flow in plants, since the distribution of individual genes can be mapped between individuals within a population and between populations. There are two phases of gene dispersal in a plant’s life cycle: pollination in which only the pollen moves and which determines how the genes will mix to form the next generation; and seed dispersal in which the new generation moves.
Gene flow is restricted in all plants, with the great majority of pollen and seeds dispersing only a short distance. This potentially divides many populations into part-separated neighborhoods, defined initially as ‘an area from which about 86% of the parents of some central individual may be treated as if drawn at random’. The most extreme restriction in pollen flow is self-fertilization where there is no genetic mixing, and after a few generations all the offspring will be genetically identical. If this is coupled with poor seed dispersal then gene flow is minimal. The opposite extreme is found in self-incompatible plants with pollen dispersed by wind; in these plants pollen can travel a long way and connect populations genetically. If these plants have an effective seed dispersal mechanism too there may be effective long distance gene flow and neighborhood size will be large. Plants pollinated by insects and vertebrates have an intermediate level of pollen dispersal, with birds, butterflies and moths dispersing pollen further than bees or flies, although these are general trends and individual species of pollinator differ in their effectiveness. At the seed dispersal stage, vertebrate-dispersed seeds travel furthest followed by wind-dispersed seeds, although many seeds have no clear dispersal mechanism beyond random events such as high winds or floods .
Natural selection
There is much debate on the relative importance of natural selection and restricted gene flow in determining how plant populations are organized genetically. Some isozymes are known to respond differently to particular environmental conditions, e.g. isozymes with different sensitivities to temperature, but in most no such differences are known and the variation has no known basis in natural selection. No selection is known that favors particular DNA sequences. This does not necessarily mean that natural selection is not important, just that we have not detected the mechanism. For all conclusions on the genetic organization of plant populations, the variation is assumed to be neutral in operation. If selection is operating, favoring one or another of the isozymes in different conditions, such as a wet or dry microhabitat in any one area, then any conclusions about gene flow must take this into account. It seems likely that many of the observed polymorphisms in plant populations are maintained in part by natural selection, since their distributions are not as predicted considering only gene flow and a relationship between certain enzyme forms and micro-environmental conditions have been found in some plants. Usually no mechanism is known involving the detected enzyme morphs but it may be a response of other linked genes or the workings of the gene in question in a
particular context within the plant.
There is evidence, from some species, that heterozygous plants with two different versions of a gene are at an advantage over homozygous plants with two genes the same. Heterozygosity normally results from a cross between two parents that are genetically distinct and frequently a self-incompatible plant will be heterozygous at many loci. In self-fertilizing plants each generation will lose, on average, half the heterozygous genes so it will rapidly become homozygous at most genetic loci. The advantage of cross-fertilization has been well known to plant breeders for centuries , known as hybrid vigor and is mainly because of increased heterozygosity. Many deleterious mutations will be masked by an alternative form and, if an enzyme occurs in two different forms, it may be able to work across a wider range of conditions. Although there is evidence from some species that heterozygotes are at an advantage, it does not appear to be universally true. Many polyploid plants, particularly those deriving initially from hybridization, can retain some heterozygosity despite self-fertilization through having two different genes from different parent species on theirhomeologous genes, i.e. the equivalent genes from the two parents. Many polyploids do self-fertilize.
Population genetic structure
The total amount of genetic variation within a species is highly variable, with species that are restricted to a small area usually much less variable than widespread species, although there are many exceptions. It is likely that the distribution of genes within a population results from a balance between natural selection and restricted gene flow. The variation that occurs can be divided into that which exists within each population and that which occurs between populations.
In some plants, the populations differ but individuals within each population vary little or not at all, a situation characteristic of self-fertilizing plants and others with restricted gene flow. In these plants restricted gene flow has the predominant effect. In others, individuals within each population vary widely but each population has a similar range of variation. This is true of plants with the potential for long-distance gene flow (see above) and particularly of many plants that dominate plant communities. In these, natural selection is likely to be much more important, and studies that have looked at gene flow directly by examining pollen and seed dispersal have consistently underestimated the extent of genetic mixing and the effective gene flow within these species. If there is strong natural selection on plants it is likely that there will be mechanisms to avoid self-fertilization since it is nearly always disadvantageous.
Those plants that spread clonally may have a ‘population’ without any variation, i.e. consisting of a single genetic individual. In most clonal species, though, there is sexual reproduction, often with a mechanism favoring cross-fertilization and, taken overall, their genetic structure may be similar to that of non-clonal species but with fewer individuals, each covering a large area.
Small populations of many plants that are isolated from any other population often have little genetic variation. This may be because of the founder effect, each being founded by one or a small number of plants at one time so have always been restricted genetically, or it may be because of genetic drift: some variants becoming extinct at random because they cannot be maintained in small populations.
Related Topics