Chapter: Essential Microbiology: Microorganisms in Genetic Engineering
Polymerase chain reaction (PCR)
Polymerase
chain reaction (PCR)
First described in the mid-1980s by Kary Mullis, PCR
is probably the most significant development in molecular biology since the
advent of gene cloning. PCR allows us to amplify a specific section of a genome
(for example a particular gene) millions of times from a tiny amount of
starting material (theoretically a single molecule!). The impact of this
powerful and highly specific process has been felt in all areas of biology and
beyond. Medicine, forensic science and evolutionary studies are but three areas
where PCR has opened up new possibilities over the last 15 years or so. To
appre- ciate how PCR works, you will need to understand the role of the enzyme
DNA polymerase in DNA replication. This is the enzyme, you’ll recall, that when
provided with single-stranded DNA and a short primer, can direct the synthesis
of a complementary second strand. Figure 12.14 illustrates the three steps in
the PCR process:
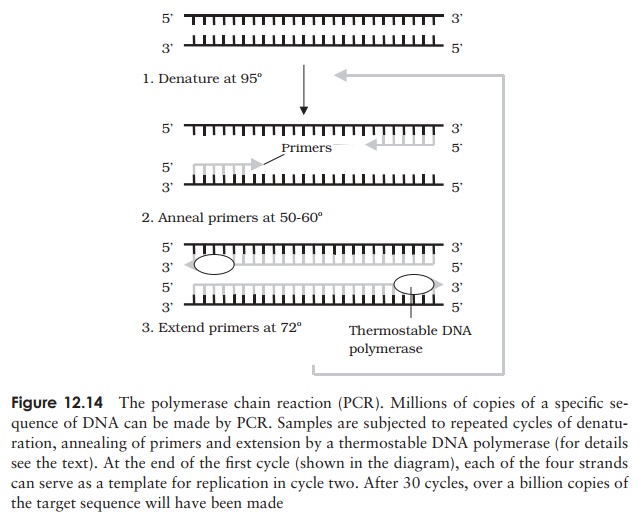
Denaturation: by heating to
95 ◦ C, the DNA is sepa rated into single strands, providing a template for
the DNA polymerase.
Primer
annealing: at a lower temperature (typically 50–60◦C, the exact
value de-pends on the primer sequence), short single-stranded primers are
allowed to attach. Primers are synthetic oligonucleotides with a sequence
complementary to the regions flanking the region we wish to amplify. One primer
anneals to each strand, at either side of the target sequence.
Polymerase
extension: at around 72◦C, the DNA
polymerase extends the primer byadding complementary nucleotides and so forms a
second strand.
The net result of this process is that instead of one
double-stranded molecule, we now have two. We now come to the key concept of
PCR; if we raise the temperature to 95 ◦ C again to
start another cycle, we will have not two, but four single-stranded templates
to work on, each of which can be converted to the double-stranded form as
before. After 20 such cycles we should have, in theory, over a million copies of our original molecule!
Typical PCR protocols run for 30–35 cycles. All this can be achieved in just a
couple of hours; the temperature cycling is carried out by a programmable
microprocessor-controlled machine called a thermal
cycler.
PCR has widespread applications in microbiology, as
in other fields of biology, but in addition, microorganisms play a crucial role
in the process itself. If you have read the above description of PCR carefully,
something should have struck you as being not quite right: how can the DNA
polymerase (a protein) tolerate being repeatedly heated to over 90 ◦ C, and what
sort of an enzyme can work effectively at 72 ◦ C? The answer
is that the DNA polymerase used in PCR comes from thermophilic bacteria such as
Thermusaquaticus, a species found
naturally in hot springs. The optimum temperature for Taq polymerase is 72 ◦ C, and it can
tolerate being raised to values considerably higher than this for short
periods. The availability of this enzyme meant that it was not necessary to add
fresh enzyme after each cycle, and the whole process could be automated, a key
factor in its subsequent phenomenal success.
Related Topics