Chapter: Biotechnology Applying the Genetic Revolution: Pathway Engineering
Polyketides and Polyketide Antibiotics
POLYKETIDES
AND POLYKETIDE ANTIBIOTICS
More and more disease-causing
microorganisms are acquiring or mutating to antibiotic resistance. This is starting
to cause problems in treating patients with certain infections.
More novel antibiotics are
becoming necessary simply to treat infections that were once susceptible to the
earlier generation of antibiotics. Although new antibiotics are still to be
found in the wild, and some may be made
by modification of a base compound (e.g., 7-ACA as described earlier), other
approaches are also useful.
Combinatorial biosynthesis
manipulates genes for useful natural products to generate a wide range of
possible variants. These can then be screened for different and/or improved
activities. In practice, only certain rather flexible biochemical pathways are
susceptible to this type of manipulation. The best example is the polyketide
pathway (Fig. 13.20). Polyketides are linear polymeric molecules. Polyketide
synthesis begins with a small initiator molecule that is elongated by addition
of subunits of two, three, or four carbons. However, each subunit contributes
two carbons to the growing polymer backbone and its other carbon atoms form
branches (methyl or ethyl groups). As
originally made, every other carbon in the polymer backbone carries a keto
group—hence the name polyketide.
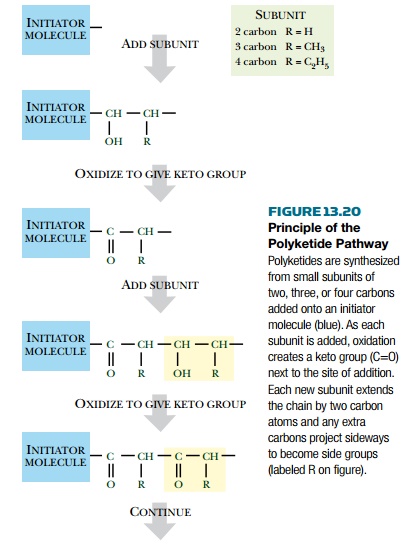
However, some of the keto
groups may be reduced to hydroxyl groups or to CH2 groups during the polymer elongation process.
After synthesis, the
polyketide chains form a variety of ring systems and are modified in various
ways. Several families of antibiotics are polyketides, the best known being the
tetracyclines and the macrolides (e.g., erythromycin).
Certain bacteria make these
antibiotics naturally, especially those of the actinomycete family. The genes
for each antibiotic pathway are usually clustered together, which is convenient
for cloning and manipulation.
The polyketide synthases
consist of modules, each of which is responsible for addition and modification
of a polyketide subunit. Each module contains activities for:
■ Loading enzyme that carries
the growing chain via a thiol ester
■ Chain extension enzyme that
selects correct incoming subunit (i.e., two-, three-, or four-carbon subunit)
and links it to the growing chain
■ Optional reductase that
converts keto to hydroxyl
■ Optional reductase that
converts hydroxyl to methylene
■ Transferase that passes
growing polyketide to the next module
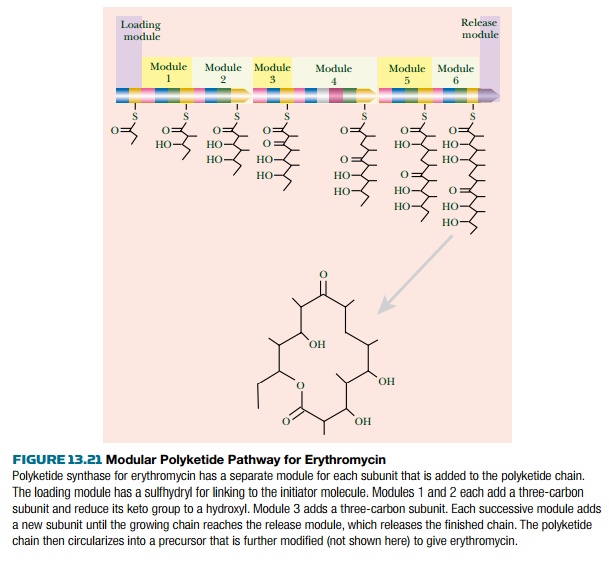
For example, the erythromycin
system has six modules, each adding a 3C subunit, plus a loading domain that picks up the initiator
molecule and a releasing domain. These
modules are arranged on three separate polypeptide chains (Fig. 13.21).
After release, the erythromycin
precursor is circularized and modified by addition of two sugar derivatives.
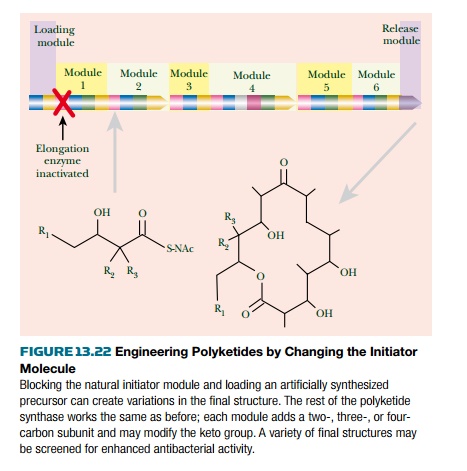
Polyketide pathways, such as
the one for erythromycin, may be engineered by two main methods. First, the
polymerizing enzyme of module 1 is inactivated. This prevents elongation of the
natural initiator molecule. An artificial analog is then added and joins the
polyketide pathway at module 2. Chemical groups present on the initiator analog
will then be present in the final polyketide product (Fig. 13.22).
An even greater variety of
polyketides can be generated by genetically altering the modules of the
polyketide synthase. For example, a module that converts the keto group to a
hydroxyl could be altered to leave the keto unchanged or to reduce it further
to a methylene (CH2) group. Or a module that incorporates a
two-carbon subunit could
be altered to use a three- or
four-carbon precursor, thus introducing methyl or ethyl branches along the
polyketide chain. For genetic engineering, the polyketide synthase gene cluster
is normally moved into E. coli. The
synthase modules may be altered by three major approaches:
(a)
Individual modules may be directly altered to inactivate or add
enzymatic activities
(b)
Modules may be replaced as a whole by other modules from the same
pathway from the same original polyketide-producing organism
(c)
Modules from the polyketide synthases of different
polyketide-producing organisms may be combined
With six successive modules, the potential for introducing variety is immense. Engineered polyketide gene clusters may be reinserted into a natural polyketide producer, such as Streptomyces, or expressed in E. coli. A library of polyketides can then be generated and screened for useful properties.
Related Topics