Chapter: Genetics and Molecular Biology: An Overview of Cell Structure and Function
Packing DNA into Cells
Packing DNA into Cells
The DNA
of the E. coli chromosome has a molecular weight of about 2 Ă— 109and thus is about 3 Ă— 106base pairs long.
Since the distancebetween base pairs in DNA is about 3.4 Ă…, the length of the
chromosome is 107 Ă… or 0.1 cm. This is very long compared to the 104
Ă… length of a bacterial cell, and the DNA must therefore wind back and forth
many times within the cell. Observation by light microscopy of living bacterial
cells and by electron microscopy of fixed and sectioned cells show, that often
the DNA is confined to a portion of the interior of the cell with dimensions
less than 0.25 µ.
To gain
some idea of the relevant dimensions, let us estimate the number of times that
the DNA of a bacterium winds back and forth within a volume we shall
approximate as a cube 0.25 µ on a
side. This will provide an idea of the average distance separating the DNA
duplexes and will also give some idea of the proportion of the DNA that lies on
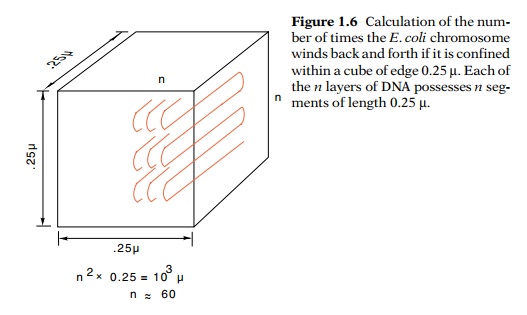
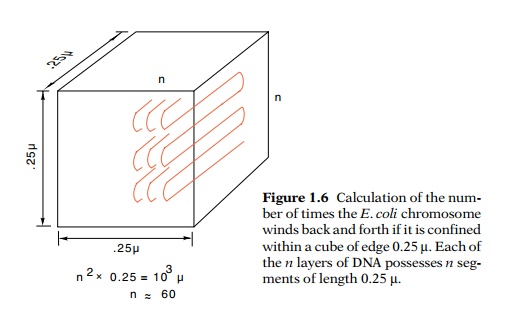
Figure
1.6 Calculation of the num-ber of
times the E. coli chromosome winds back and forth if it is confined within a
cube of edge 0.25 µ. Each of the n
layers of DNA possesses n seg-ments
of length 0.25 µ.
the surface of the chromosomal mass. The number of
times, N, that the DNA must wind back
and forth will then be related to the length of the DNA and the volume in which
it is contained. If we approximate the path of the DNA as consisting of
n layers, each layer consisting of n segments of length 0.25 µ (Fig. 1.6), the total number of
segments is n2. Therefore,
2,500n2 Ă… = 107
Ă… and n = 60. The spacing between
adjacent segments of the DNA is 2,500 Ă…/60 = 40 Ă….
The close
spacing between DNA duplexes raises the interesting prob-lem of accessibility
of the DNA. RNA polymerase has a diameter of about 100 Ă… and it may not fit
between the duplexes. Therefore, quite possibly only DNA on the surface of the
nuclear mass is accessible for transcrip-tion. On the other hand, transcription
of the lactose and arabinose operons can be induced within as short a time as
two seconds after adding inducers. Consequently either the nuclear mass is in
such rapid motion that any portion of the DNA finds its way to the surface at
least once every several seconds, or the RNA polymerase molecules do penetrate
to the interior of the nuclear mass and are able to begin transcription of any
gene at any time. Possibly, start points of the arabinose and lactose operons
always reside on the surface of the DNA.
Compaction
of the DNA generates even greater problems in eu-karyotic cells. Not only do
they contain up to 1,000 times the amount of the DNA found in bacteria, but the
presence of the histones on the DNA appears to hinder access of RNA polymerase
and other enzymes to the DNA. In part, this problem is solved by regulatory
proteins binding to regulatory regions before nucleosomes can form in these
positions. Apparently, upon activation of a gene additional regulatory proteins
bind, displacing more histones, and transcription begins. The DNA of many
eukaryotic cells is specially contracted before cell division, and at this time
it actually does become inaccessible to RNA polymerase. At all times, however,
accessibility of the DNA to RNA polymerase must be hindered.
Related Topics