Chapter: Genetics and Molecular Biology: Advanced Genetic Engineering
Finding Clones from a Known Amino Acid Sequence
Finding Clones from a Known Amino Acid Sequence
Sometimes the protein product of a gene is
available in pure form. This happy circumstance can be used to facilitate
cloning of the gene. Portions of the protein can be sequenced to determine a
potential DNA sequence that could have encoded this portion of the protein. An
oligonucleotide with this sequence can then be used to screen a collec-tion of
clones, which is called a library, to detect those containing complementary
sequences. The screening is done as described. Occasionally, a clone is found
in the libraries which hybridizes to the screening oligonucleotide, but which
is not the correct clone. This results from the chance occurrence of a sequence
comple-mentary to the probing oligonucleotide. These incorrect positives can be
detected by screening with a second oligonucleotide that should hybridize to a
different part of the gene encoding the protein in question. Only the desired
clones should hybridize to both oligonucleotides.
The redundancy in the genetic code prevents simple
reverse transla-tion from an amino acid sequence to a DNA sequence. The
difficulty caused by the redundancy can be partially overcome by using portions
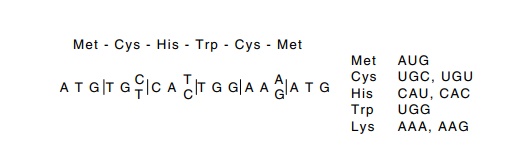
Figure
10.1 Reverse translating to obtain the
sequences that could have en-coded a short peptide.
of the protein’s sequence containing amino acids
whose codon redun-dancy is low. This is possible since both tryptophan and
methionine have unique codons. Consider the sequence met-cys-his-trp-lys-met.
Only one codon specifies an internal methionine, while the cysteine, histidine,
and lysine are each specified by only two possible codons. Therefore one of
only 1 × 2 × 2 × 1 × 2 × 1 = 8 sequences encoded the six amino acids (Fig.
10.1).
The eight necessary oligonucleotides can be
synthesized simultane-ously by machine by incorporating either of the two
ambiguous nucleo-tides at the necessary positions. This is accomplished simply
by supplying at the correct time a mixture of the two nucleotides to the
synthesis solution.
Purification
of the protein necessary for the oligonucleotide probing approach often is
straightforward. Conventional purification need not be performed, however.
Since all that is needed for the cloning is determination of portions of the
amino acid sequence, purification and detection methods need not preserve the
protein’s native structure. SDS gel electrophoresis, for example, can be used
as a final step in the purification of the protein. The protein in the correct
band in the gel can be eluted and a portion of its amino terminal sequence
determined by gas phase and mass spectrometry. As little as 10 -12
moles of protein are sufficient for determining enough of the sequence that
oligonu-cleotide probes can be designed
to identify clones carrying the gene.
Related Topics