Chapter: Biochemistry: Protein Synthesis: Translation of the Genetic Message
Eukaryotic Translation
Eukaryotic Translation
The main
features of translation are the same in prokaryotes and eukaryotes, but the
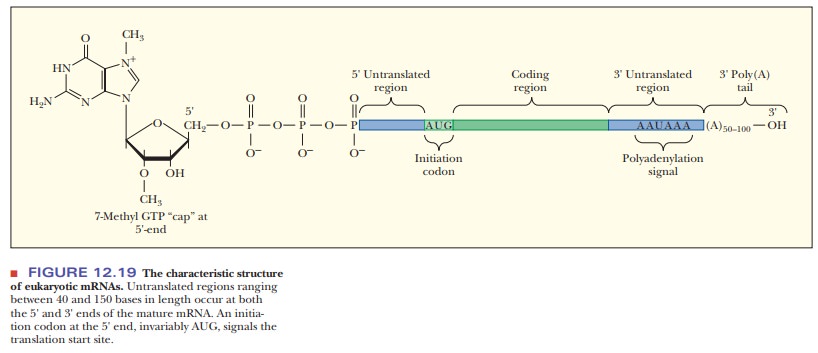
details differ. The messenger RNAs of eukaryotes are characterized by two major
posttranscriptional modifications. The first is the 5' cap, and the second is
the 3' poly A tail (Figure 12.19). Both modifications are essential to
eukaryotic translation.
How is translation different in eukaryotes?
Chain Initiation
This is
the part of eukaryotic translation that is the most different from that in
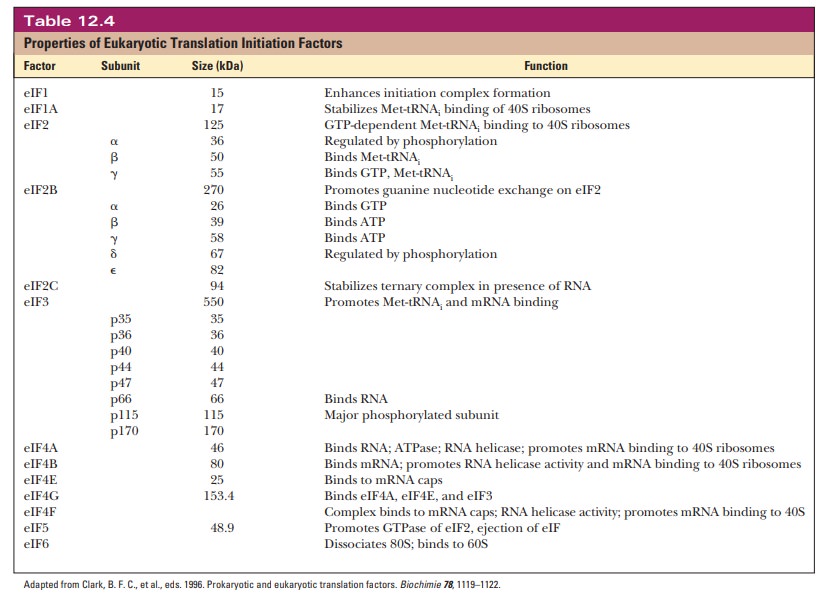
prokaryotes. Thirteen more initiation factors are given the designation eIF, for eukaryotic initiation factor. Many of them are multisubunit
proteins. Table 12.4summarizes pertinent information about these initiation factors.
Step 1 in chain initiation involves the assembly of a 43S preinitiation com-plex (Figure 12.20). The initial amino acid is methionine, which is attached to a special tRNAi that serves only as the initiator tRNA. There is no fmet in eukaryotes. The met-tRNAi is delivered to the 40S ribosomal subunit as a complex with GTP and eIF2. The 40S ribosome is also bound to eIF1A and eIF3. This order of events is different from that in prokaryotes in that the first tRNA binds to the ribosome without the presence of the mRNA. In Step 2, the mRNA is recruited. There is no Shine–Dalgarno sequence for location of the start codon. The 5' cap orients the ribosome to the correct AUG via what is called a scanning mechanism, which is driven by ATP hydrolysis. The eIF4E is also a cap-binding protein, which forms a complex with several other eIFs. A poly A binding protein (Pab1p) links the poly A tail to eIF4G. The eIF-40S complex is initially positioned upstream of the start codon (Figure 12.21).
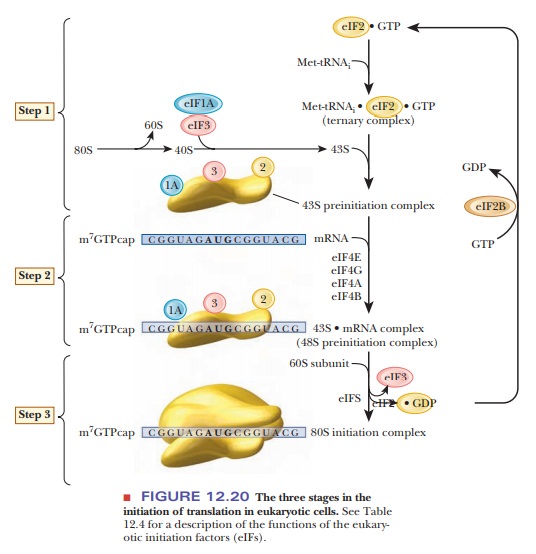
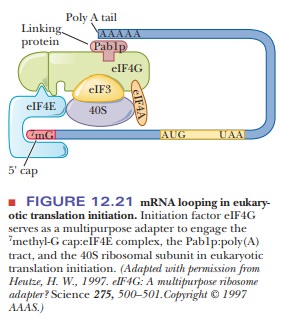
It moves downstream until it encounters the first AUG in the correct con-text. The context is determined by a few bases surrounding the start codon, called the Kozak sequence. It is characterized by the consensus sequence –3ACCAUGG+4. The ribosome may skip the first AUG it finds if the next one has the Kozak sequence. Another factor is the presence of mRNA secondary structure.
If hairpin loops form downstream of an AUG, an earlier AUG may be chosen. The
mRNA and the seven eIFs constitute the 48S preinitiation complex. In Step 3,
the 60S ribosome is recruited, forming the 80S initiation complex. GTP is
hydrolyzed, and the initiation factors are released.
Chain Elongation
Peptide
chain elongation in eukaryotes is very similar to that of prokaryotes. The same
mechanism of peptidyl transferase and ribosome translocation is seen. The
structure of the eukaryotic ribosome is different in that there is no E site,
only the A and P sites. There are two eukaryotic elongation factors, eEF1 and
eEF2. The eEF1 consists of two subunits, eEF1A and eEF1B. The 1A subunit is the
counterpart of EF-Tu in prokaryotes. The 1B subunit is the equivalent of the
EF-Ts in prokaryotes. The eEF2 protein is the counterpart of the prokaryotic
EF-G, which causes translocation.
Many of
the differences between translation in prokaryotes and eukaryotes can be seen
in the response to inhibitors of protein synthesis and to toxins. The
antibiotic chloramphenicol (a trade name is Chloromycetin) binds to the A site
and inhibits peptidyl transferase activity in prokaryotes, but not in eukaryotes.
This property has made chloramphenicol useful in treating bacterial
infec-tions. In eukaryotes, diphtheria toxin is a protein that interferes with
protein synthesis by decreasing the activity of the eukaryotic elongation
factor eEF2.
Chain Termination
As in
prokaryotic termination, the ribosome encounters a stop codon, either UAG, UAA,
or UGA, and these are not recognized by a tRNA molecule. In prokaryotes, three
different release factors-RF1, RF2, and RF3-were used, with two of them
alternating, depending on which stop codon was found. In eukaryotes, only one
release factor binds to all three stop codons and catalyzes the hydrolysis of
the bond between the C-terminal amino acid and the tRNA.
There is
a special tRNA called a suppressor tRNA,
which allows translation to continue through a stop codon. Suppressor tRNAs
tend to be found in cells in which a mutation has introduced a stop codon.
Coupled Transcription and Translation in Eukaryotes?
Until
recently, the dogma of eukaryotic translation was that it was physically
separated from transcription. Transcription occurred in the nucleus, and mRNA
was then exported to the cytosol for translation. Although this system is
accepted as the normal process, recent evidence has shown that the nucleus has
all of the components (mRNA, ribosomes, protein factors) necessary for
translation. In addition, evidence shows that, in isolated test systems,
proteins are translated in the nucleus. The authors of the most recent work
suggest that 10%–15% of the cell’s protein synthesis occurs in the nucleus.
Summary
Eukaryotic translation involves many more
protein factors than the cor-responding translation in prokaryotes.
Both the 5' cap and the 3'
poly A tail are involved in orienting the ribo-some close to the correct AUG
used as the start codon. There is no Shine–Dalgarno sequence in eukaryotic
mRNA.
Once bound, the ribosome
moves down the mRNA scanning for the correct AUG until it finds one that is in the
correct context, which is identified by a small mRNA sequence around the AUG
called a Kozak sequence.
Eukaryotic chain elongation
is similar to the prokaryotic counterpart. With chain termination, there is
only one release factor that binds to all three stop codons.
It has
recently been found that there is some coupled transcription and translation in
the nucleus of eukaryotic cells.
Related Topics