Chapter: Biotechnology Applying the Genetic Revolution: Protein Engineering
DNA Shuffling
DNA
SHUFFLING
Natural selection works on
new sequences generated both by mutation and recombination. DNA shuffling is a method of artificial
evolution that includes the creation of novel mutations as well as recombination. The gene to be improved is cut
into random segments around 100 to 300 base pairs long. The segments are then
reassembled by using a suitable DNA polymerase with overlapping segments or by
using some version of overlap PCR. This recombines segments from different
copies of the same gene (Fig. 11.10).
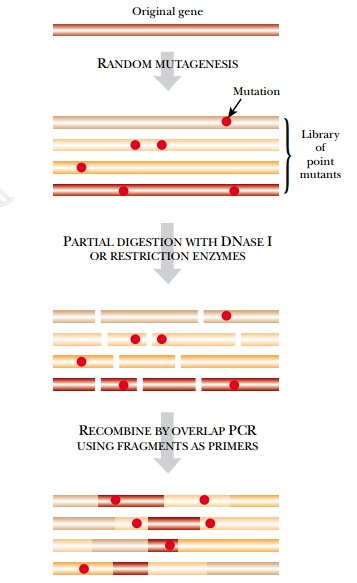

Mutations may be introduced
in several ways, including the standard mutagenesis procedures already
described. In addition, the DNA segments may be generated using error-prone PCR
(see earlier discussion) instead of by using restriction enzymes.
Alternatively, mutations may be introduced during the reassembly procedure
itself by using a DNA polymerase that has impaired proofreading capability. The
result is a large number of copies of the gene, each with several mutations
scattered at random throughout its sequence. The final shuffled and mutated
gene copies must then be expressed and screened for altered properties of the
encoded protein.
A more powerful variant of
DNA shuffling is to start with several closely related (i.e., homologous)
versions of the same gene from different organisms. The genes are cut at random
with appropriate restriction enzymes and the segments mixed before reassembly.
The result is a mixture of genes that have recombined different segments from
different original genes
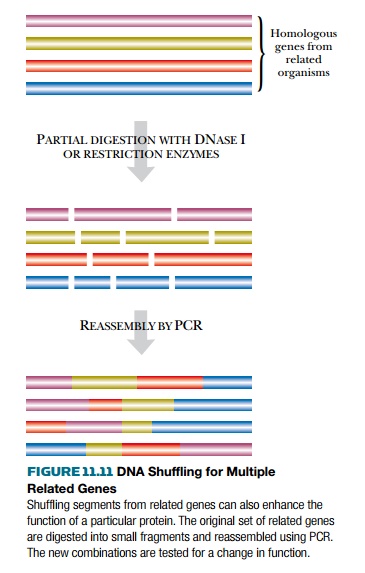
(Fig. 11.11). Note that the
reassembled segments keep their original natural order. For example, several
related β-lactamases from different
enteric bacteria have been shuffled. The shuffled genes were cloned onto a
plasmid vector and transformed into host bacteria. The bacteria were then
screened for resistance to selected β-lactam antibiotics. This approach yielded
improved β-lactamases that degraded
certain penicillins and cephalosporins more rapidly and so made their host cells up to 500-fold more resistant
to these β-lactam antibiotics.
Related Topics