Botany : Chromosomal Basis of Inheritance - DNA Metabolism in Plants | 12th Botany : Chapter 3 : Chromosomal Basis of Inheritance
Chapter: 12th Botany : Chapter 3 : Chromosomal Basis of Inheritance
DNA Metabolism in Plants
DNA
Metabolism in Plants
As the repository of
genetic information, DNA occupies a unique and central place among biological
macromolecules. The structure of DNA is a marvelous device for the storage of
genetic information. The term “DNA Metabolism” can be used to describe process
by which copies of DNA molecules are made (replication) along with repair and
recombination.
In this chapter we
briefly discuss about the DNA metabolism in plants
DNA Replication: In the double helix the
two parental strands of DNA separate and each parental strand synthesizes a
new complementary strand. DNA replication is semiconservative, i.e each new DNA
molecule conserves one original strand.
DNA Repair: How is genomic stability
maintained in all living organisms? How do organisms on earth survive? What
is essential for their survival?
DNA is unique because it
is the only macromolecule where the repair system exists, which recognises and
removes mutations. DNA is subjected to various types of damaging reactions such
as spontaneous or environmental agents or natural endogenous threats. Such
damages are corrected by repair enzymes and proteins, immediately after the
damage has taken place. DNA repair system plays a major role in maintaining the
genomic / genetic integrity of the organism. DNA repair systems protect the
integrity of genomes from genotoxic stresses.
Recombination: In cells the genetic information
within and among DNA molecule are re-arranged by a process called genetic
recombination. Recombination is the result of crossing over between the pairs
of homologous chromosomes during meiosis. In earlier classes you have learnt
chromosomal recombination. In molecular level it involves breakage and reunion
of polynucleotides.
1. Eukaryotic DNA replication
Replication starts at a
specific site on a DNA sequence known as the Origin of replication. There are
more than one origin of replication in eukaryotes. Saccharomyces
cerevisiae (yeast) has approximately 400 origins of replication. DNA replication
in eukaryotes starts with the assembly of a prereplication complex (preRC)
consisting of 14 different proteins. Part of a preRC is a group of 6 proteins
called the origin recognition complex (ORC) which acts as initiator in
eukaryotic DNA replication. The origin of replication in yeast is called
as ARS sites (Autonomously Replicating Sequences). In yeast,
ORC was identified as a protein complex which binds directly to ARS elements.
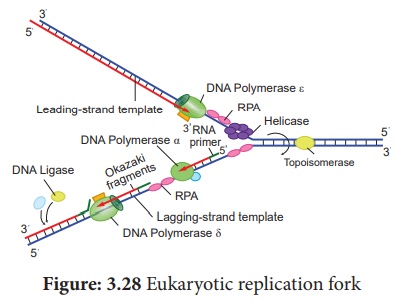
Replication fork is the
site (point of unwinding) of separation of parental DNA strands where new
daughter strands are formed. Multiple replication forks are found in
eukaryotes. The enzyme helicases are involved in unwinding of DNA by
breaking hydrogen bonds holding the two strands of DNA and replication protein
A (RPA) prevents the separated polynucleotide strand from getting reattached.
Topoisomerase is an enzyme which breaks
DNAs covalent bonds and removes positive supercoiling ahead of replication
fork. It eliminates the torsional stress caused by unwinding of DNA double
helix.
DNA replication is
initiated by an enzyme DNA polymerase α / primase which synthesizes short
stretch of RNA primers on both leading strand (continuous DNA strand) and
lagging strands (discontinuous DNA strand). Primers are needed because DNA
polymerase requires a free 3’ OH to initiate synthesis. DNA polymerase
covalently connects the nucleotides at the growing end of the new DNA strand.
DNA Pol α (alpha), DNA
Pol δ (delta) and DNA Pol ε (Epsilon) are the 3 enzymes involved in nuclear DNA
replication.
DNA Pol α – Synthesizes short
primers of RNA
DNA Pol δ – Main Replicating enzyme
of cell nucleus
DNA Pol ε – Extend the DNA Strands
in replication fork
DNA Polymerase β does
not play any role in the replication of normal DNA. Function Removing incorrect bases from damaged DNA. It
is involved in Base excision repair.
DNA Synthesis takes place in 5’ → 3’ direction and it is semidiscontinuous. When DNA is synthesized in 5’ → 3’ direction, only in the free 3’ end (OH end) DNA is elongated. In 1960s Reiji Okazaki and his colleagues found out that one of the new DNA strands is synthesized in short pieces called Okazaki fragments. In discontinuous strand where the Okazaki fragments are united by ligase is called Lagging strand where the replication direction is 5’ → 3’ which is opposite to the direction of fork movement.
The continuous strand is
called Leading strand where the replication direction is 5’ -> 3’ which is
same to the direction to that of the replication fork movement. DNA ligase
joins any nicks in the DNA by forming a phosphodiester bond between 3’ hydroxyl
and 5’ phosphate group.
Arabidopsis telomere sequence - TTTAGGG
Plants Lacks Telomere Clock
Plant meristematic cells
produces telomerase so the meristematic cells has an unlimited ability to
divide. You have already studied about the telomeres in Chapter 6 and 8 of
Class XI. In plants telomeres do not shrink as in somatic cells of vertebrates.
Telomerase levels are higher in root tips and seedlings (renewable tissue) which
has a higher amount of meristematic cells than proliferative structures like
leaves.
What is the special mechanism which replicates chromosomal ends?
After the replication of
the chromosomes, the enzyme telomerase adds several more repeats of DNA
sequences to the telomeres. Telomerase use short RNA molecules as a template
and add repeat sequences on to telomeres (DNA nucleotide polymerisation).
The Energetics of DNA
Replication - Deoxyribonucleotides such as deoxyadenosine triphosphate dATP,
dGTP, dCTP and dTTP provide energy for the synthesis of DNA. Purpose of
Deoxyribonucelotides (1) acts as a substrate (2) provide energy for
polymerisation.
2. Experimental evidence of DNA replication: Taylors Experiment
J. Herbert Taylor,
Philip Woods and Walter Hughes demonstrated the semiconservative replication of
DNA in the root cells of Vicia faba. They labelled DNA with
3H Thymidine, a radioactive precursor of DNA and
performed autoradiography. They grew root tips in a medium in the presence of
radioactive labelled thymidine, so that the radioactivity was incorporated into
the DNA of these cells. The outline of this labelled chromosomes appears in the
form of scattered black dots of silver grains on a photographic film.
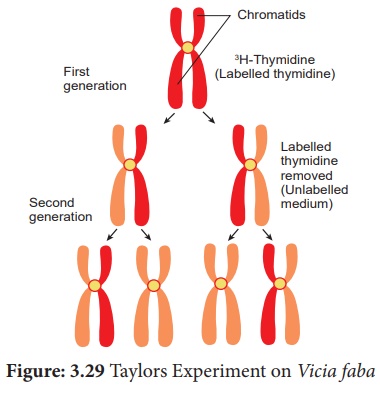
The root tips with labelled chromosomes were placed in an unlabelled medium containing colchicine to arrest the culture at the metaphase and examine the chromosome by autoradiography. The observations were,
1. In the chromosome of first
generation the radioactivity was found to be distributed to both
the chromatids because in the original strand of DNA double helix
was labelled with radioactivitiy and the new strand was unlabelled.
2. In the chromosome of
the second generation only one of the two chromatids in each chromosome
was radioactive (labelled).
The results proved the
semiconservative method of DNA replication.
Related Topics