Chapter: Genetics and Molecular Biology: Genetic Engineering and Recombinant DNA
Cutting DNA with Restriction Enzymes
Cutting DNA with Restriction Enzymes
The restriction enzymes provide a necessary tool
for cutting fragments of DNA out of larger molecules. Their exquisite
specificity permits very great selectivity, and because more than one hundred
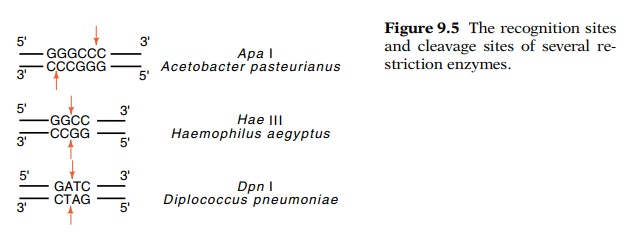
different restriction enzymes are known (Fig. 9.5), their wide variety permits
much choice in the cleavage sites utilized. Often, fragments may be produced
with end points located within 20 base pairs of any desired location.
One of the more useful properties of restriction
enzymes for genetic engineering is found in the restriction-modification system
produced by the E. coli plasmid R. The corresponding
restriction enzyme is called EcoRI.
Instead of cleaving at the center of its palindromic recognitionsequence, this
enzyme cleaves off-center and produces four base self-complementary ends. These
“sticky” ends are most useful in recombi
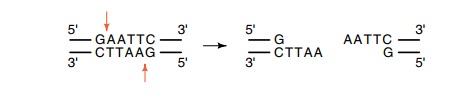
This permits efficient
joining of DNA fragments during ligation steps. About half of the restriction
enzymes now known generate overhanging or sticky ends. In some situations, a
DNA fragment can even be arranged to have two different types of sticky ends so
that its insertion into another DNA can be forced to proceed in one particular
orientation.
Related Topics