Chapter: Genetics and Molecular Biology: Advanced Genetic Engineering
Chromosome Mapping
Chromosome Mapping
It is simplest first to determine a large-scale and
coarse map of a chromosome and then to build upon the initial map to increase
resolution. Construction of a low-resolution physical map can begin with a
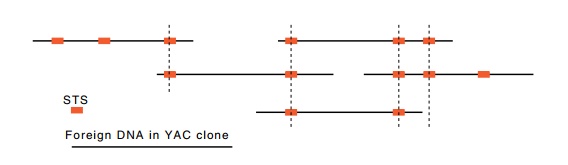
Figure
10.11 A set of DNA fragments ordered by
the STS sites they share incommon.
library of overlapping
clones. Mapping would then be picking and ordering a subset of these which
spans an entire chromosome. The larger the clones, the fewer which need to be
ordered, and the easier it is to construct of the initial map. The yeast
artificial chromosome vectors, YAC vectors, are useful for chromosome mapping
projects because they can accommodate very large fragments of foreign DNA, up
to 106 base pairs. These contain autonomous replicating sequences,
ARS, as origins and telomeres for the ends.
Let us
first consider the physical markers which are necessary for the construction of
a physical map. Unique sequences of DNA one or two hundred base pairs long can
provide a good starting point for construc-tion of a physical map. Such
sequences can be obtained by sequencing random clones made from the chromosome
to be mapped. The unique-ness of a sequence can be checked by hybridization.
From such a unique sequence, two 20 to 30 base portions can be chosen for
oligonucleotide primers that will permit PCR amplification of the sequence
lying be-tween. This amplification will occur only if the template DNA present
in the PCR reaction contains the unique sequence. Such a unique
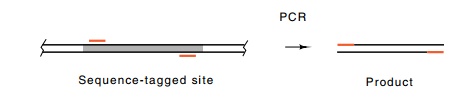
sequence is called a sequence-tagged site, or STS.
A sample of DNA is tested for the presence of an STS by PCR amplification using
the two oligonucleotides followed by gel electrophoresis to assay for the
ampli-fication of the proper-sized DNA fragment.
When the number of sequence-tagged sites exceeds the number of clones necessary to span the chromosome by a factor of four or five or more, the sites can be used just like restriction enzyme cleavage sites are used in a chromosomal walk. That is, each YAC vector is assayed for the presence of each STS. If an STS site is present in two vectors, then the chromosome fragment carried by each contains this region in common (Fig. 10.11). Merely from this data, the STS sites can be ordered and an overlapping physical map of the chromosome can be constructed. Of course, for even the smallest human chromosome, tens of thousands of YAC vectors must be present in the library, and hun-dreds of STS sites are needed. Therefore, very many PCR amplification reactions will be required for the ordering.
Mutations,
insertions, and deletions have also been used in chromo-some mapping.
Originally, restriction fragment length polymorphisms, RFLPs were used as
genetic and physical markers for chromosome mapping. Over the majority of the
genome, sequence differences be-tween individuals exist at a frequency of about
10-4. If one of these differences occurs in the cleavage site for a
restriction enzyme an RFLP will exist there. Another source of an RFLP is an
insertion or deletion or a difference in the number of repeats of a short
sequence contained between the two restriction enzyme cleavage sites.
Restriction
fragment length polymorphisms can be detected using Southern transfers as
described in an earlier section. If individuals differ in one out of 104
base pairs, then the chances that a particular restriction enzyme cleavage site
will be present in one chromosome but be missing in the homologous chromosome
would be about 0.001. There is a reasonably high probability that either an
insertion exists between two restriction enzyme cleavage sites or that two
individuals differ at a particular point in the number of times that a short
sequence is repeated there. One out of a hundred to one out of a thousand
restriction fragments it is likely to be polymorphic between some individuals
in the population. Therefore, considerable work could be required to find DNA
sequences useful for RFLP mapping. One method for RFLP detection is to clone
random segments of human DNA and use them to probe Southern transfers of
various enzyme digests from a moderate number of people. The occasional clone
which reveals a polymorphism is then mapped to a specific chromosome and
finally mapped at higher resolu-tion by comparing its segregation pattern to
the pattern of other markers known to lie on the same chromosome.
A more
efficient method of finding and mapping genetic markers utilizes the polymerase
chain reaction. Instead of using Southern trans-fers to detect an RFLP, PCR is
used to amplify a segment of DNA that contains variable numbers of inserts. Different individuals in the popu-
Figure
10.12 A set of DNA fragments ordered by
the STS sites they share incommon.
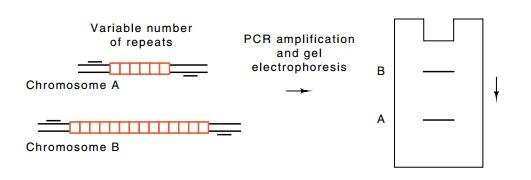
lation often possess different numbers of inserts
at the same point (Fig. 10.12). The size of a fragment synthesized by PCR
identifies the size of the insert. It is much easier to perform a PCR
amplification and determine the size of the product than it is to perform a
Southern transfer. Therefore, most chromosome mapping will shift to the use of
PCR. By the approaches described above, hundreds of RFLP or PCR-based markers
have already been mapped to the human genome and DNA defects of a significant
number of genetic diseases have been mapped with respect to the markers.
Related Topics