Chapter: Biochemistry: Nucleic Acid Metabolism
Biosynthesis of DNA
Biosynthesis of DNA
The process by which the new double helical DNA
synthesized from the existing DNA is called as "Replication". The
mechanism of biosynthesis of DNA has been largely clarified by the discovery of
the enzyme DNA Polymerase or DNA Nucleotidyl transferase. This enzyme catalyses
the polymerization of mononucleotides to polynucleotides, which needs the
following for its action.
1.
A
template strand dictates the synthesis of the new daughter strand and sequence
of the template strand determines the addition of the nucleotides.
2.
RNA
primer to which subsequent nucleotides can be added.
3.
Four
nucleoside triphosphates namely, dGTP, dATP, dTTP and dCTP.
4.
Magnesium
ions as co-factor.
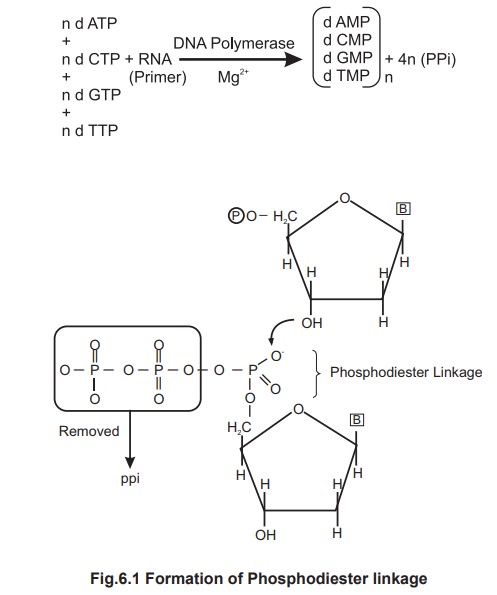
The energy required for this reaction is
provided by the hydrolysis of high energy bonds in the linear triphosphate
units of the dATP, dCTP, dGTP & dTTP. As each monomer is incorporated into
new chain, it loses its terminal pyrophosphate unit (PPi).
1. Replication
It is a process in which DNA copies itself to
produce identical daughter molecules of DNA.
Models of Replication
Three models of replication had been proposed.
They are 1. Conservative replication 2.Dispersive replication and 3.Semi
conservative replication.
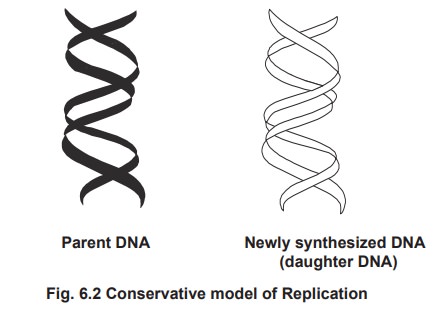
1. Conservative replication
According to this model, the parental DNA is
conserved to one daughter cell and the newly synthesized DNA to another
daughter cell.
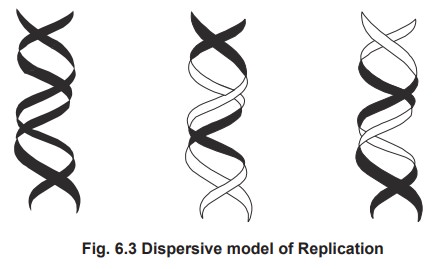
2. Dispersive Replication
According to this model, the parental DNA is
unequally distributed (randomly) to the daughter cells.
3. Semiconservative Replication
Semiconseravative model of replication showing
the daughter DNA having one parental strand and one daughter strand.
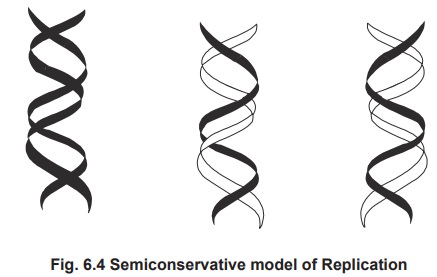
This model was established to be the correct
mode of replication by the experiment carried out by Messlson and Stahl in
1957. E. coli cells were grown in a
medium containing 15NH4Cl, for many generations such that the E.coli
cells have density labelled 15N atoms in their DNA. The cells were then grown
in a medium containing unlabelled NH4Cl. DNA was harvested from the
daughter cells after one generation and the density of the DNA was analyzed
using Cesium chloride density gradient centrifugation. If replication happens
to occur by conservative model, two bands are corresponding to heavy DNA and
other unlabelled DNA should be got. But if replication is semi-conservative,
daughter DNAs should possess one labelled strand and one unlabelled strand, so
they will form only one band with intermediate density. The results show that
they form bands with intermediate density alone in the first generation, which
confirms the semiconservative model of replication.
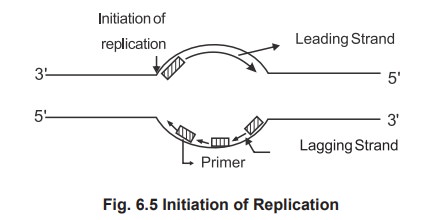
2. Sequential Process of Replication Initiation of DNA replication
o Initiation of DNA synthesis occurs at a site
called origin of replication.
o In prokaryotes, only one origin.
o In eukaryotes, there are multiple origins.
o The origin consists of a short sequence of A = T
base pairs.
Replication bubbles
The two complementary strands of DNA separate at
a site of replication to form a bubble. In eukaryotes many replication bubbles
occur.
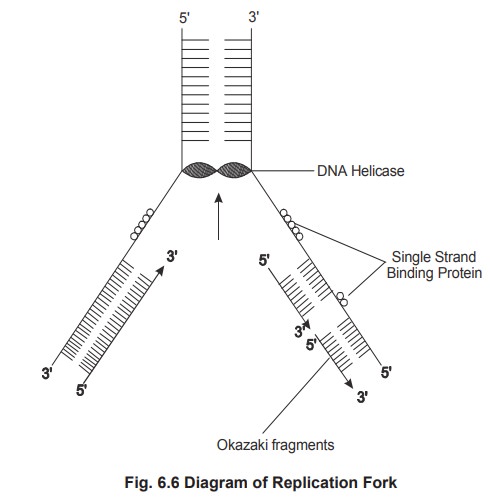
Unwinding of parental DNA
After the initiation point is located the
unwinding of the DNA takes place once in every 10 nucleotide pairs. This allows
the “strand separation”.
In prokaryotes the unwinding of the structure
occurs with the help of an enzyme called helicase, which requires energy of
hydrolysis of two ATP molecules per base pair broken. Another protein single
strand binding protein (SSB), binds to the unwound DNA to prevent (rewinding)
rejoining.
As the DNA polymerase cannot initiate
replication a primer is a needed and the primer is the small nucleotide of RNA,
synthesised by enzymes primase.
Under the influence of DNA polymerase, in the
presence of Mg2+, the double strands of the DNA acting as a template (or
primer) separate by cleaving the hydrogen bonds between complementary bases.
The deoxyribo nucleoside triphosphates are attracted from solution in the
cellular sap to form hydrogen bonds with their complementary bases on the
separated strands of the (primer RNA) template dictates the sequence in which
the monomers are assembled.
Polymerisation
During this reaction, each incoming nucleotide
loses a pyrophosphate group and forms an ester linkage with the 3' hydroxyl
group of the deoxyribose on the existing last nucleotide. This linkage is
called “phosphodiester linkage”.
The parental strands run in antiparallel
direction. Synthesis occur simultaneously on both strands, but at different
rates. No enzymes can synthesize 3' ® 5' direction and a single enzyme can not
synthesize both strands. The single enzyme replicates one strand called leading
strand in a continuous manner in 5' to 3' direction (forward), it replicates
the other strand, lagging strand in a discontinuous manner and polymerising
only few (250) nucleotides again run in 5' to 3' at backward direction. This is
called semicontinuous DNA synthesis. The newly synthesized DNA is made as
discontinuous small fragments called as Okazaki fragments and joined by the
enzyme called ligase.
Thus 2 daughter double helices are formed each
consisting of an old strand of the primer DNA and a complementary new strand.
The final composition and nucleotide sequence of each strand is identical with
the corresponding strand in the template (parent) DNA. This process has been
named as replication.
Related Topics